Gene
KWMTBOMO16230
Pre Gene Modal
BGIBMGA013981
Annotation
PREDICTED:_protein_Wnt-2-like_[Bombyx_mori]
Full name
Protein Wnt
+ More
Protein Wnt-7b
Protein Wnt-7b
Location in the cell
Extracellular Reliability : 4.256
Sequence
CDS
ATGTGTCGGTCTTCTCCATCGGCTATAGCCGCAATCGGTGATGGACTTAAGATGGCATATGAAGAATGCCGGGCTCAATTGAGCGGCCACAGGTGGAACTGCTCAGGAGTGGGAGACGGAAATCACTTCGGGCACGTTATGCCGTTAGCAACGCGTGAGGCGGCTTTTACGTACGCCATCACATCGGCGGGGGTCACCCATGCTTTGAGCATGGCCTGCGCTCGTGGGGAGCTGCCTGGTTGTGGATGTACAGGAAATAAGAGGCGTACGATAAACACAATCGAGCAGTTCCAATGGGGCGGATGCGGTTCCAGTGCGTACGGCGCTAGGTTAGCCCGGCGGTTTTTGGACGCGAGAGAATTGGAACGAGACGGCAGAACCCTCATGAACCTCCACAACAACCGGGTCGGAAGGAAGGTGAGCGAGGTGGTAAAGGACCTAGTTCATCGCGAGTGCAAATGCCACGGCGTCTCGGGCTCCTGCGCTATGAGGACTTGCTGGCGGGCGCTACCGCAGTTCAGGGCTGCGGCTGCCACCCTGCGGGACAAGTATAAGCGAGCGAAGCTAGTTATGCCGCACCCAGCAACGAATCCACAGGACATTAACGTTCGAACACACCTCGTAATAAAACGACAAAAGCACCCAGGTCTCGGTCGACAGCCCAGGAAATCGGAGCTGGTGTTTCTCGAACAGTCACCTTCCTACTGCGAGCCTGATACTGCAGCAGGATCGTTTGGAACTCACGGAAGGATATGCAATAGGACGTCAAGAGGTGAGGACGGCTGCGAGACCCTATGCTGCGGACGCGGGTACAGCACGACCCGCAGCGTTGAGGAGACAAAGTGCCGCTGCCAGTTCCACTGGTGCTGCAACGTCACGTGCAGCAAATGCGTGACGACAGTGGAAACGCACCTTTGTAATTAG
Protein
MCRSSPSAIAAIGDGLKMAYEECRAQLSGHRWNCSGVGDGNHFGHVMPLATREAAFTYAITSAGVTHALSMACARGELPGCGCTGNKRRTINTIEQFQWGGCGSSAYGARLARRFLDARELERDGRTLMNLHNNRVGRKVSEVVKDLVHRECKCHGVSGSCAMRTCWRALPQFRAAAATLRDKYKRAKLVMPHPATNPQDINVRTHLVIKRQKHPGLGRQPRKSELVFLEQSPSYCEPDTAAGSFGTHGRICNRTSRGEDGCETLCCGRGYSTTRSVEETKCRCQFHWCCNVTCSKCVTTVETHLCN
Summary
Description
Ligand for members of the frizzled family of seven transmembrane receptors.
Ligand for members of the frizzled family of seven transmembrane receptors that functions in the canonical Wnt/beta-catenin signaling pathway (PubMed:15923619, PubMed:28803732). Required for normal fusion of the chorion and the allantois during placenta development (PubMed:11543617). Required for central nervous system (CNS) angiogenesis and blood-brain barrier regulation (PubMed:28803732).
Ligand for members of the frizzled family of seven transmembrane receptors that functions in the canonical Wnt/beta-catenin signaling pathway (PubMed:30026314). Required for normal fusion of the chorion and the allantois during placenta development (By similarity). Required for central nervous system (CNS) angiogenesis and blood-brain barrier regulation (PubMed:30026314).
Ligand for members of the frizzled family of seven transmembrane receptors that functions in the canonical Wnt/beta-catenin signaling pathway (PubMed:15923619, PubMed:28803732). Required for normal fusion of the chorion and the allantois during placenta development (PubMed:11543617). Required for central nervous system (CNS) angiogenesis and blood-brain barrier regulation (PubMed:28803732).
Ligand for members of the frizzled family of seven transmembrane receptors that functions in the canonical Wnt/beta-catenin signaling pathway (PubMed:30026314). Required for normal fusion of the chorion and the allantois during placenta development (By similarity). Required for central nervous system (CNS) angiogenesis and blood-brain barrier regulation (PubMed:30026314).
Subunit
Forms a soluble 1:1 complex with AFM; this prevents oligomerization and is required for prolonged biological activity. The complex with AFM may represent the physiological form in body fluids (By similarity). Interacts with FZD1 and FZD10 (PubMed:15923619). Interacts with FZD4 (in vitro) (PubMed:15923619). Interacts with PORCN (PubMed:10866835). Interacts with glypican GPC3 (By similarity). Interacts (via intrinsically disordered linker region) with RECK; interaction with RECK confers ligand selectivity for Wnt7 in brain endothelial cells and allows these cells to selectively respond to Wnt7 (By similarity).
Forms a soluble 1:1 complex with AFM; this prevents oligomerization and is required for prolonged biological activity (PubMed:16227623). The complex with AFM may represent the physiological form in body fluids (PubMed:26902720). Interacts with FZD1 and FZD10. Interacts with FZD4 (in vitro). Interacts with PORCN (By similarity). Interacts with glypican GPC3 (PubMed:16227623). Interacts (via intrinsically disordered linker region) with RECK; interaction with RECK confers ligand selectivity for Wnt7 in brain endothelial cells and allows these cells to selectively respond to Wnt7 (PubMed:30026314).
Forms a soluble 1:1 complex with AFM; this prevents oligomerization and is required for prolonged biological activity (PubMed:16227623). The complex with AFM may represent the physiological form in body fluids (PubMed:26902720). Interacts with FZD1 and FZD10. Interacts with FZD4 (in vitro). Interacts with PORCN (By similarity). Interacts with glypican GPC3 (PubMed:16227623). Interacts (via intrinsically disordered linker region) with RECK; interaction with RECK confers ligand selectivity for Wnt7 in brain endothelial cells and allows these cells to selectively respond to Wnt7 (PubMed:30026314).
Similarity
Belongs to the Wnt family.
Keywords
Complete proteome
Developmental protein
Disulfide bond
Extracellular matrix
Glycoprotein
Lipoprotein
Reference proteome
Secreted
Signal
Wnt signaling pathway
Feature
chain Protein Wnt
Uniprot
H9JWR7
A0A0N1I6U7
A0A212FE41
A0A0N0P9S5
A0A2H1V4K0
A0A1Y1MUT8
+ More
A0A1Y1MRN2 A0A1Y1MWA3 A0A1Y1MV11 D6WT32 A0A1B6CTV4 K7IUK3 A0A1Q3FR18 A0A1Q3FQZ0 B0XE42 A0A3S1B792 A0A210PIC4 A0A1S4FJ50 Q16Z87 A0A1W4WQL4 A0A1W7R6K2 A0A182J8Y0 Q75PH8 A0A2K5SA58 Q75PH6 A0A2K5SA60 A0A0K8VCL5 A0A2K5SA53 A0A2K5SA56 A0A2R8NBC2 A0A2U3TVB0 U3BHR4 A0A2K6BRD2 A0A2K6MDG1 A0A2K5VJG3 A0A2K6RQ32 A0A2K5HJB1 A0A0P6J925 G5BVA8 A0A2I3GKQ7 A0A2R9BAW7 A0A2K6BRK5 A0A2K6MD94 A0A2I2YNF6 A0A2R9BAW6 A0A2I3LY45 A0A2K5P8L3 A0A2J8TW64 A0A2K5HJ68 G1RTL0 A0A2J8LMI4 A8K0G1 A0A2I2ZE89 V3Z3G3 I3NCV6 A0A286Y3S2 B8A595 A0A2J8TW35 H0VRH8 A0A1U7U421 F7HJM7 A0A2I2Y7P3 A0A0G2K3B7 Q6NZR1 A0A2K6BRC8 A0A2K6MDF6 A0A2K5VJ85 A0A2I3MYX8 A0A2K6RQ28 A0A2K5P8M7 A0A2K5HJ48 A0A1D5R7Q0 A0A2I3HGF4 H0VZV3 Q5NSW6 P28047 E9PU66 A0A2K6BRE8 A0A2K5VJD2 A0A2I3HZL1 A0A2K5ZBV8 A0A0D9QZ47 A0A096NKR7 A0A2K5P8S6 H2P4S1 A0A2I3TH72 P56706 I3N7Q1 A0A2K6RQ41 A0A2K6MDF5 A0A2K6RQ24 A0A2K5HJD1 A0A2R9BEM4 A0A1D5R803 A0A2K5P8M5 A0A1B0GBR3 G3ICH8 A0A2K5ZBW6 A0A1U7Q3Z5 D2HKK9
A0A1Y1MRN2 A0A1Y1MWA3 A0A1Y1MV11 D6WT32 A0A1B6CTV4 K7IUK3 A0A1Q3FR18 A0A1Q3FQZ0 B0XE42 A0A3S1B792 A0A210PIC4 A0A1S4FJ50 Q16Z87 A0A1W4WQL4 A0A1W7R6K2 A0A182J8Y0 Q75PH8 A0A2K5SA58 Q75PH6 A0A2K5SA60 A0A0K8VCL5 A0A2K5SA53 A0A2K5SA56 A0A2R8NBC2 A0A2U3TVB0 U3BHR4 A0A2K6BRD2 A0A2K6MDG1 A0A2K5VJG3 A0A2K6RQ32 A0A2K5HJB1 A0A0P6J925 G5BVA8 A0A2I3GKQ7 A0A2R9BAW7 A0A2K6BRK5 A0A2K6MD94 A0A2I2YNF6 A0A2R9BAW6 A0A2I3LY45 A0A2K5P8L3 A0A2J8TW64 A0A2K5HJ68 G1RTL0 A0A2J8LMI4 A8K0G1 A0A2I2ZE89 V3Z3G3 I3NCV6 A0A286Y3S2 B8A595 A0A2J8TW35 H0VRH8 A0A1U7U421 F7HJM7 A0A2I2Y7P3 A0A0G2K3B7 Q6NZR1 A0A2K6BRC8 A0A2K6MDF6 A0A2K5VJ85 A0A2I3MYX8 A0A2K6RQ28 A0A2K5P8M7 A0A2K5HJ48 A0A1D5R7Q0 A0A2I3HGF4 H0VZV3 Q5NSW6 P28047 E9PU66 A0A2K6BRE8 A0A2K5VJD2 A0A2I3HZL1 A0A2K5ZBV8 A0A0D9QZ47 A0A096NKR7 A0A2K5P8S6 H2P4S1 A0A2I3TH72 P56706 I3N7Q1 A0A2K6RQ41 A0A2K6MDF5 A0A2K6RQ24 A0A2K5HJD1 A0A2R9BEM4 A0A1D5R803 A0A2K5P8M5 A0A1B0GBR3 G3ICH8 A0A2K5ZBW6 A0A1U7Q3Z5 D2HKK9
Pubmed
19121390
26354079
22118469
28004739
18362917
19820115
+ More
20075255 28812685 17510324 25243066 25362486 21993625 22722832 22398555 15164055 16136131 10591208 23254933 21993624 17431167 15057822 15489334 19468303 12147135 2279700 10866835 11543617 15923619 28803732 11562755 8168088 16227623 26902720 30026314 21804562 29704459 20010809
20075255 28812685 17510324 25243066 25362486 21993625 22722832 22398555 15164055 16136131 10591208 23254933 21993624 17431167 15057822 15489334 19468303 12147135 2279700 10866835 11543617 15923619 28803732 11562755 8168088 16227623 26902720 30026314 21804562 29704459 20010809
EMBL
BABH01018551
KQ460689
KPJ12754.1
AGBW02008980
OWR51990.1
KQ459166
+ More
KPJ03538.1 ODYU01000645 SOQ35763.1 GEZM01023608 JAV88280.1 GEZM01023607 JAV88281.1 GEZM01023611 JAV88276.1 GEZM01023612 JAV88275.1 KQ971354 EFA07553.2 GEDC01020533 JAS16765.1 AAZX01004215 AAZX01005813 GFDL01005051 JAV29994.1 GFDL01005004 JAV30041.1 DS232802 EDS45875.1 RQTK01000562 RUS77655.1 NEDP02076657 OWF36235.1 CH477495 EAT39973.1 GEHC01000851 JAV46794.1 AB167809 BAD12587.1 AB167811 BAD12589.1 GDHF01032883 GDHF01015620 JAI19431.1 JAI36694.1 GAMT01006051 GAMT01006050 GAMT01006049 GAMT01006048 GAMT01006047 GAMT01006046 JAB05812.1 AQIA01006583 AQIA01006584 AQIA01006585 AQIA01006586 GEBF01003593 JAO00040.1 JH172084 EHB13222.1 ADFV01129020 ADFV01129021 ADFV01129022 ADFV01129023 ADFV01129024 ADFV01129025 ADFV01129026 ADFV01129027 ADFV01129028 ADFV01129029 AJFE02039433 AJFE02039434 AJFE02039435 CABD030121578 CABD030121579 CABD030121580 AHZZ02001400 NDHI03003480 PNJ37239.1 AACZ04043605 NBAG03000283 PNI48474.1 BX511035 CR536603 AK289526 BAF82215.1 KB203301 ESO85168.1 AGTP01080758 AGTP01080759 AGTP01080760 AGTP01080761 AGTP01080762 AGTP01080763 AGTP01080764 AGTP01080765 AGTP01080766 AGTP01080767 AAKN02054985 AAKN02054986 PNJ37237.1 JSUE03005760 JSUE03005761 JSUE03005762 AABR07058588 AABR07073335 AC112534 AC115763 BC066003 AAH66003.1 AB197137 BAD83363.1 M89802 BC052018 AQIB01150601 AQIB01150602 ABGA01134833 ABGA01134834 ABGA01220113 ABGA01220114 ABGA01220115 ABGA01220116 ABGA01220117 ABGA01220118 ABGA01220119 ABGA01220120 PNJ37235.1 PNI48470.1 AB062766 AF416743 BC034923 CCAG010005876 CCAG010005877 CCAG010005878 CCAG010005879 CCAG010005880 JH001927 RAZU01000074 EGV98910.1 RLQ75186.1 GL192962 EFB24521.1
KPJ03538.1 ODYU01000645 SOQ35763.1 GEZM01023608 JAV88280.1 GEZM01023607 JAV88281.1 GEZM01023611 JAV88276.1 GEZM01023612 JAV88275.1 KQ971354 EFA07553.2 GEDC01020533 JAS16765.1 AAZX01004215 AAZX01005813 GFDL01005051 JAV29994.1 GFDL01005004 JAV30041.1 DS232802 EDS45875.1 RQTK01000562 RUS77655.1 NEDP02076657 OWF36235.1 CH477495 EAT39973.1 GEHC01000851 JAV46794.1 AB167809 BAD12587.1 AB167811 BAD12589.1 GDHF01032883 GDHF01015620 JAI19431.1 JAI36694.1 GAMT01006051 GAMT01006050 GAMT01006049 GAMT01006048 GAMT01006047 GAMT01006046 JAB05812.1 AQIA01006583 AQIA01006584 AQIA01006585 AQIA01006586 GEBF01003593 JAO00040.1 JH172084 EHB13222.1 ADFV01129020 ADFV01129021 ADFV01129022 ADFV01129023 ADFV01129024 ADFV01129025 ADFV01129026 ADFV01129027 ADFV01129028 ADFV01129029 AJFE02039433 AJFE02039434 AJFE02039435 CABD030121578 CABD030121579 CABD030121580 AHZZ02001400 NDHI03003480 PNJ37239.1 AACZ04043605 NBAG03000283 PNI48474.1 BX511035 CR536603 AK289526 BAF82215.1 KB203301 ESO85168.1 AGTP01080758 AGTP01080759 AGTP01080760 AGTP01080761 AGTP01080762 AGTP01080763 AGTP01080764 AGTP01080765 AGTP01080766 AGTP01080767 AAKN02054985 AAKN02054986 PNJ37237.1 JSUE03005760 JSUE03005761 JSUE03005762 AABR07058588 AABR07073335 AC112534 AC115763 BC066003 AAH66003.1 AB197137 BAD83363.1 M89802 BC052018 AQIB01150601 AQIB01150602 ABGA01134833 ABGA01134834 ABGA01220113 ABGA01220114 ABGA01220115 ABGA01220116 ABGA01220117 ABGA01220118 ABGA01220119 ABGA01220120 PNJ37235.1 PNI48470.1 AB062766 AF416743 BC034923 CCAG010005876 CCAG010005877 CCAG010005878 CCAG010005879 CCAG010005880 JH001927 RAZU01000074 EGV98910.1 RLQ75186.1 GL192962 EFB24521.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000053268
UP000007266
UP000002358
+ More
UP000002320 UP000271974 UP000242188 UP000008820 UP000192223 UP000075880 UP000233040 UP000008225 UP000233120 UP000233180 UP000233100 UP000233200 UP000233080 UP000006813 UP000001073 UP000240080 UP000001519 UP000028761 UP000233060 UP000002277 UP000005640 UP000030746 UP000005215 UP000005447 UP000189704 UP000006718 UP000002494 UP000000589 UP000233140 UP000029965 UP000001595 UP000092444 UP000001075 UP000273346 UP000189706
UP000002320 UP000271974 UP000242188 UP000008820 UP000192223 UP000075880 UP000233040 UP000008225 UP000233120 UP000233180 UP000233100 UP000233200 UP000233080 UP000006813 UP000001073 UP000240080 UP000001519 UP000028761 UP000233060 UP000002277 UP000005640 UP000030746 UP000005215 UP000005447 UP000189704 UP000006718 UP000002494 UP000000589 UP000233140 UP000029965 UP000001595 UP000092444 UP000001075 UP000273346 UP000189706
Pfam
PF00110 wnt
ProteinModelPortal
H9JWR7
A0A0N1I6U7
A0A212FE41
A0A0N0P9S5
A0A2H1V4K0
A0A1Y1MUT8
+ More
A0A1Y1MRN2 A0A1Y1MWA3 A0A1Y1MV11 D6WT32 A0A1B6CTV4 K7IUK3 A0A1Q3FR18 A0A1Q3FQZ0 B0XE42 A0A3S1B792 A0A210PIC4 A0A1S4FJ50 Q16Z87 A0A1W4WQL4 A0A1W7R6K2 A0A182J8Y0 Q75PH8 A0A2K5SA58 Q75PH6 A0A2K5SA60 A0A0K8VCL5 A0A2K5SA53 A0A2K5SA56 A0A2R8NBC2 A0A2U3TVB0 U3BHR4 A0A2K6BRD2 A0A2K6MDG1 A0A2K5VJG3 A0A2K6RQ32 A0A2K5HJB1 A0A0P6J925 G5BVA8 A0A2I3GKQ7 A0A2R9BAW7 A0A2K6BRK5 A0A2K6MD94 A0A2I2YNF6 A0A2R9BAW6 A0A2I3LY45 A0A2K5P8L3 A0A2J8TW64 A0A2K5HJ68 G1RTL0 A0A2J8LMI4 A8K0G1 A0A2I2ZE89 V3Z3G3 I3NCV6 A0A286Y3S2 B8A595 A0A2J8TW35 H0VRH8 A0A1U7U421 F7HJM7 A0A2I2Y7P3 A0A0G2K3B7 Q6NZR1 A0A2K6BRC8 A0A2K6MDF6 A0A2K5VJ85 A0A2I3MYX8 A0A2K6RQ28 A0A2K5P8M7 A0A2K5HJ48 A0A1D5R7Q0 A0A2I3HGF4 H0VZV3 Q5NSW6 P28047 E9PU66 A0A2K6BRE8 A0A2K5VJD2 A0A2I3HZL1 A0A2K5ZBV8 A0A0D9QZ47 A0A096NKR7 A0A2K5P8S6 H2P4S1 A0A2I3TH72 P56706 I3N7Q1 A0A2K6RQ41 A0A2K6MDF5 A0A2K6RQ24 A0A2K5HJD1 A0A2R9BEM4 A0A1D5R803 A0A2K5P8M5 A0A1B0GBR3 G3ICH8 A0A2K5ZBW6 A0A1U7Q3Z5 D2HKK9
A0A1Y1MRN2 A0A1Y1MWA3 A0A1Y1MV11 D6WT32 A0A1B6CTV4 K7IUK3 A0A1Q3FR18 A0A1Q3FQZ0 B0XE42 A0A3S1B792 A0A210PIC4 A0A1S4FJ50 Q16Z87 A0A1W4WQL4 A0A1W7R6K2 A0A182J8Y0 Q75PH8 A0A2K5SA58 Q75PH6 A0A2K5SA60 A0A0K8VCL5 A0A2K5SA53 A0A2K5SA56 A0A2R8NBC2 A0A2U3TVB0 U3BHR4 A0A2K6BRD2 A0A2K6MDG1 A0A2K5VJG3 A0A2K6RQ32 A0A2K5HJB1 A0A0P6J925 G5BVA8 A0A2I3GKQ7 A0A2R9BAW7 A0A2K6BRK5 A0A2K6MD94 A0A2I2YNF6 A0A2R9BAW6 A0A2I3LY45 A0A2K5P8L3 A0A2J8TW64 A0A2K5HJ68 G1RTL0 A0A2J8LMI4 A8K0G1 A0A2I2ZE89 V3Z3G3 I3NCV6 A0A286Y3S2 B8A595 A0A2J8TW35 H0VRH8 A0A1U7U421 F7HJM7 A0A2I2Y7P3 A0A0G2K3B7 Q6NZR1 A0A2K6BRC8 A0A2K6MDF6 A0A2K5VJ85 A0A2I3MYX8 A0A2K6RQ28 A0A2K5P8M7 A0A2K5HJ48 A0A1D5R7Q0 A0A2I3HGF4 H0VZV3 Q5NSW6 P28047 E9PU66 A0A2K6BRE8 A0A2K5VJD2 A0A2I3HZL1 A0A2K5ZBV8 A0A0D9QZ47 A0A096NKR7 A0A2K5P8S6 H2P4S1 A0A2I3TH72 P56706 I3N7Q1 A0A2K6RQ41 A0A2K6MDF5 A0A2K6RQ24 A0A2K5HJD1 A0A2R9BEM4 A0A1D5R803 A0A2K5P8M5 A0A1B0GBR3 G3ICH8 A0A2K5ZBW6 A0A1U7Q3Z5 D2HKK9
PDB
6AHY
E-value=7.51534e-61,
Score=591
Ontologies
KEGG
PATHWAY
GO
GO:0016055
GO:0007275
GO:0005576
GO:0005102
GO:0030182
GO:0005109
GO:0005615
GO:0045165
GO:0060070
GO:0071300
GO:0072089
GO:0048144
GO:0048018
GO:0061180
GO:0021871
GO:0032536
GO:0005886
GO:0007257
GO:0005737
GO:0048812
GO:0031175
GO:0051384
GO:0046330
GO:0072054
GO:0045879
GO:0032364
GO:0010628
GO:0060710
GO:0016332
GO:0048568
GO:0001701
GO:0022009
GO:0008284
GO:0003338
GO:0060482
GO:0072205
GO:0072236
GO:1902262
GO:0060033
GO:0072053
GO:0045669
GO:0060669
GO:0072207
GO:0060535
GO:0050768
GO:1902379
GO:0050808
GO:0001525
GO:0051145
GO:0042592
GO:0044237
GO:0060484
GO:0001944
GO:0072061
GO:0048864
GO:0036516
GO:1904938
GO:0005788
GO:0072060
GO:0060425
GO:0060428
GO:0030324
GO:0060560
GO:0021846
GO:0009986
GO:0042475
GO:0030666
GO:0070062
GO:0005796
GO:0070307
GO:0008233
GO:0001678
GO:0005536
GO:0046835
GO:0006281
GO:0006259
PANTHER
Topology
Subcellular location
Secreted
Extracellular space
Extracellular matrix
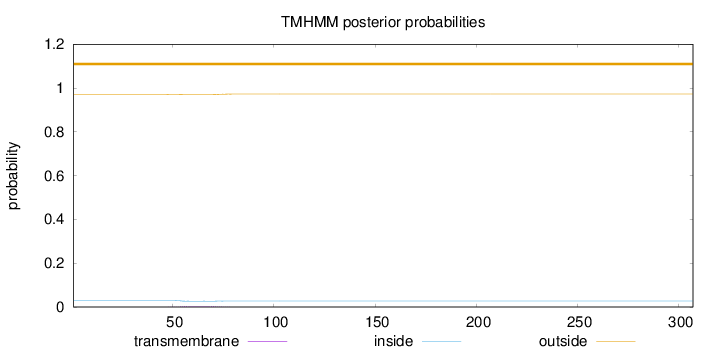
Length:
307
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0798199999999999
Exp number, first 60 AAs:
0.02929
Total prob of N-in:
0.02978
outside
1 - 307
Population Genetic Test Statistics
Pi
212.551584
Theta
153.878494
Tajima's D
1.600066
CLR
0.180158
CSRT
0.809859507024649
Interpretation
Uncertain
