Gene
KWMTBOMO16211 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013916
Annotation
PREDICTED:_6-phosphogluconolactonase_isoform_X1_[Bombyx_mori]
Full name
6-phosphogluconolactonase
Location in the cell
Cytoplasmic Reliability : 2.977
Sequence
CDS
ATGACGATCATCAAAGTTATTGATGAAGAAGAAATCATCAGAAAACTATCGACATACATTCAAAAGATTTCAAACGACGCGATCTTGAATAGAAATAAATTCGTAGTTGGACTTTCTGGCGGCTCTGTCGTCAAATATTTATGTGAAGGTTTACCACAAGTTGAAACAGACTGGTCTAAATGGACTCTAGCCTTCTGTGATGAAAGAGTAGTACCTGAAGACAGCAGCGATTCTACATTCGGAATATACAAGAAAGATTTAATCCCTAAGACTGAATTAAAGGAGAGCCAGTTTATAACAATCAAACAAGGAGCTACAGCTCAAGAAACAGCAAAGGATTACATTGAAAAGTTAAGAAAAGTCTTTGGTGGAGATGATTTCAAATTTGACTTACTTCTACTTGGCATGGGACCAGATGGACACACATGCTCCCTGTTCCCAGGTCACAAATTGCTTGAAGAAACAGAAGATAAGGTAGCAGCTATAACAGATTCCCCAAAACCTCCACCGGAGAGAATCACATTAACTTATCCAGTTATAAATGGAGCAAGGAACTGTATTTTTGCTATTTCTGGGGCAGGGAAGTCAGAAATGGCTAAACGTATTCTAAAAGACAAAGAAGATTTACCAGCTGGTCGAGTGAAGCCGACAGACGGCTCTCTATATTGGATCCTGGACGAAGAAGCAGCTGTACATTTGTAA
Protein
MTIIKVIDEEEIIRKLSTYIQKISNDAILNRNKFVVGLSGGSVVKYLCEGLPQVETDWSKWTLAFCDERVVPEDSSDSTFGIYKKDLIPKTELKESQFITIKQGATAQETAKDYIEKLRKVFGGDDFKFDLLLLGMGPDGHTCSLFPGHKLLEETEDKVAAITDSPKPPPERITLTYPVINGARNCIFAISGAGKSEMAKRILKDKEDLPAGRVKPTDGSLYWILDEEAAVHL
Summary
Description
Hydrolysis of 6-phosphogluconolactone to 6-phosphogluconate.
Catalytic Activity
6-phospho-D-glucono-1,5-lactone + H2O = 6-phospho-D-gluconate + H(+)
Similarity
Belongs to the glucosamine/galactosamine-6-phosphate isomerase family. 6-phosphogluconolactonase subfamily.
Uniprot
H9JWK2
A2TF14
A0A0N1INN4
A0A194Q017
A0A212FC63
S4P9E4
+ More
A0A2H1WMK7 A0A0L7LDU1 A0A2A4JCA4 A0A1W4XRF1 A0A2W1BG55 A0A2P8XFX3 A0A0C9Q5I1 D6WWZ6 A0A1B6CM29 A0A336MWD1 A0A1L8E2G0 A0A1B6H374 A0A232FBU0 K7IW11 A0A336MJQ0 A0A1Y1NEX0 A0A1L8E1T4 A0A0P4W4X3 A0A1L8E1Z4 A0A1B6KH60 A0A0P5KH27 A0A0P5BVL0 A0A0P5YXZ1 A0A154NZ42 A0A0N8DM66 A0A1L8E1M6 A0A310SHD3 A0A0P6IGJ5 A0A146LI57 A0A0A9WAB3 A0A0P5U6P0 A0A1I8N1L4 W8BW14 U4UTG0 A0A0K8SQJ7 N6U308 A0A0N0BL12 A0A1I8QA94 A0A2R7VY42 E2A481 A0A0L7QXE0 A0A069DQY8 F4WMX9 A0A224XJM6 A0A0P5CSK3 A0A195FUK7 A0A151HYV3 A0A158NSK5 A0A1S3KGS3 A0A023F1S0 A0A1B6K9M7 A0A0V0G8Q8 T1HIG0 A0A087TTH8 A0A151WIL5 A0A0A1XPE9 A0A2A3ESK3 A0A1B0GKZ1 A0A0P5E676 A0A0L0BMF2 A0A0P5T1I7 A0A0N7Z8V6 A0A210Q6A3 A0A0P5RE35 A0A0K8UDB3 W4XGH6 A0A026W4R4 T1JA20 A0A088A588 A0A2C9K2K9 A0A034V1A5 V4B8D7 E9GDE3 A0A0P5TED1 A0A1I8MXD4 A0A0B7B4I1 A0A023GGW8 G3MPZ5 U5EWT6 A0A131XJM1 A0A2S2PLM6 A0A067RLS3 A0A2H8THQ6 A0A0J7KT26 J9K0I9 A0A293MPE1 H3AHH1 A0A1D1W620 A0A1J1HM86 A0A224YS02 A0A131YTI9 L7M3L3 A0A131Y354 A0A0P5CEK3
A0A2H1WMK7 A0A0L7LDU1 A0A2A4JCA4 A0A1W4XRF1 A0A2W1BG55 A0A2P8XFX3 A0A0C9Q5I1 D6WWZ6 A0A1B6CM29 A0A336MWD1 A0A1L8E2G0 A0A1B6H374 A0A232FBU0 K7IW11 A0A336MJQ0 A0A1Y1NEX0 A0A1L8E1T4 A0A0P4W4X3 A0A1L8E1Z4 A0A1B6KH60 A0A0P5KH27 A0A0P5BVL0 A0A0P5YXZ1 A0A154NZ42 A0A0N8DM66 A0A1L8E1M6 A0A310SHD3 A0A0P6IGJ5 A0A146LI57 A0A0A9WAB3 A0A0P5U6P0 A0A1I8N1L4 W8BW14 U4UTG0 A0A0K8SQJ7 N6U308 A0A0N0BL12 A0A1I8QA94 A0A2R7VY42 E2A481 A0A0L7QXE0 A0A069DQY8 F4WMX9 A0A224XJM6 A0A0P5CSK3 A0A195FUK7 A0A151HYV3 A0A158NSK5 A0A1S3KGS3 A0A023F1S0 A0A1B6K9M7 A0A0V0G8Q8 T1HIG0 A0A087TTH8 A0A151WIL5 A0A0A1XPE9 A0A2A3ESK3 A0A1B0GKZ1 A0A0P5E676 A0A0L0BMF2 A0A0P5T1I7 A0A0N7Z8V6 A0A210Q6A3 A0A0P5RE35 A0A0K8UDB3 W4XGH6 A0A026W4R4 T1JA20 A0A088A588 A0A2C9K2K9 A0A034V1A5 V4B8D7 E9GDE3 A0A0P5TED1 A0A1I8MXD4 A0A0B7B4I1 A0A023GGW8 G3MPZ5 U5EWT6 A0A131XJM1 A0A2S2PLM6 A0A067RLS3 A0A2H8THQ6 A0A0J7KT26 J9K0I9 A0A293MPE1 H3AHH1 A0A1D1W620 A0A1J1HM86 A0A224YS02 A0A131YTI9 L7M3L3 A0A131Y354 A0A0P5CEK3
EC Number
3.1.1.31
Pubmed
19121390
26354079
22118469
23622113
26227816
28756777
+ More
29403074 18362917 19820115 28648823 20075255 28004739 26823975 25401762 25315136 24495485 23537049 20798317 26334808 21719571 21347285 25474469 25830018 26108605 27129103 28812685 24508170 15562597 25348373 23254933 21292972 22216098 28049606 24845553 9215903 27649274 28797301 26830274 25576852
29403074 18362917 19820115 28648823 20075255 28004739 26823975 25401762 25315136 24495485 23537049 20798317 26334808 21719571 21347285 25474469 25830018 26108605 27129103 28812685 24508170 15562597 25348373 23254933 21292972 22216098 28049606 24845553 9215903 27649274 28797301 26830274 25576852
EMBL
BABH01018477
EF198104
ABM90638.1
KQ460955
KPJ10244.1
KQ459583
+ More
KPI98633.1 AGBW02009220 OWR51320.1 GAIX01009100 JAA83460.1 ODYU01009334 SOQ53684.1 JTDY01001611 KOB73366.1 NWSH01002132 PCG69070.1 KZ150509 PZC70643.1 PYGN01002284 PSN30910.1 GBYB01009318 JAG79085.1 KQ971361 EFA08071.1 GEDC01022863 JAS14435.1 UFQT01002057 SSX32557.1 GFDF01001389 JAV12695.1 GECZ01000634 JAS69135.1 NNAY01000450 OXU28304.1 UFQT01001350 SSX30160.1 GEZM01007487 GEZM01007486 GEZM01007485 GEZM01007484 GEZM01007483 JAV95155.1 GFDF01001391 JAV12693.1 GDRN01101384 JAI58391.1 GFDF01001390 JAV12694.1 GEBQ01029194 JAT10783.1 GDIQ01191076 JAK60649.1 GDIP01183501 JAJ39901.1 GDIP01053576 JAM50139.1 KQ434783 KZC04842.1 GDIP01020119 JAM83596.1 GFDF01001451 JAV12633.1 KQ759893 OAD62087.1 GDIQ01011478 JAN83259.1 GDHC01011440 JAQ07189.1 GBHO01040161 GBHO01013253 JAG03443.1 JAG30351.1 GDIP01117261 JAL86453.1 GAMC01013161 GAMC01013158 GAMC01013155 JAB93394.1 KB632356 ERL93410.1 GBRD01010182 JAG55642.1 APGK01041544 KB740994 ENN75955.1 KQ435687 KOX81368.1 KK854152 PTY12249.1 GL436596 EFN71752.1 KQ414705 KOC63226.1 GBGD01002822 JAC86067.1 GL888223 EGI64449.1 GFTR01003739 JAW12687.1 GDIP01171703 JAJ51699.1 KQ981261 KYN44101.1 KQ976720 KYM76708.1 ADTU01025002 ADTU01025003 ADTU01025004 ADTU01025005 GBBI01003255 JAC15457.1 GEBQ01031852 JAT08125.1 GECL01001774 JAP04350.1 ACPB03010283 KK116673 KFM68417.1 KQ983089 KYQ47651.1 GBXI01001542 JAD12750.1 KZ288191 PBC34494.1 AJWK01033970 GDIP01146504 GDIQ01198925 JAJ76898.1 JAK52800.1 JRES01001654 KNC21113.1 GDIP01132363 JAL71351.1 GDKW01002283 JAI54312.1 NEDP02004840 OWF44258.1 GDIQ01121552 JAL30174.1 GDHF01027761 JAI24553.1 AAGJ04037077 AAGJ04037078 KK107419 EZA51070.1 JH431980 GAKP01022678 JAC36274.1 KB203357 ESO84989.1 GL732540 EFX82067.1 GDIP01127664 JAL76050.1 HACG01040330 HACG01040331 CEK87195.1 CEK87196.1 GBBM01002304 JAC33114.1 JO843946 AEO35563.1 GANO01002822 JAB57049.1 GEFH01002261 JAP66320.1 GGMR01017157 MBY29776.1 KK852601 KDR20495.1 GFXV01001858 MBW13663.1 LBMM01003488 KMQ93476.1 ABLF02031720 GFWV01022810 MAA47537.1 AFYH01136520 AFYH01136521 BDGG01000019 GAV08881.1 CVRI01000004 CRK87566.1 GFPF01005608 MAA16754.1 GEDV01006702 JAP81855.1 GACK01006113 JAA58921.1 GEFM01002858 JAP72938.1 GDIP01171704 JAJ51698.1
KPI98633.1 AGBW02009220 OWR51320.1 GAIX01009100 JAA83460.1 ODYU01009334 SOQ53684.1 JTDY01001611 KOB73366.1 NWSH01002132 PCG69070.1 KZ150509 PZC70643.1 PYGN01002284 PSN30910.1 GBYB01009318 JAG79085.1 KQ971361 EFA08071.1 GEDC01022863 JAS14435.1 UFQT01002057 SSX32557.1 GFDF01001389 JAV12695.1 GECZ01000634 JAS69135.1 NNAY01000450 OXU28304.1 UFQT01001350 SSX30160.1 GEZM01007487 GEZM01007486 GEZM01007485 GEZM01007484 GEZM01007483 JAV95155.1 GFDF01001391 JAV12693.1 GDRN01101384 JAI58391.1 GFDF01001390 JAV12694.1 GEBQ01029194 JAT10783.1 GDIQ01191076 JAK60649.1 GDIP01183501 JAJ39901.1 GDIP01053576 JAM50139.1 KQ434783 KZC04842.1 GDIP01020119 JAM83596.1 GFDF01001451 JAV12633.1 KQ759893 OAD62087.1 GDIQ01011478 JAN83259.1 GDHC01011440 JAQ07189.1 GBHO01040161 GBHO01013253 JAG03443.1 JAG30351.1 GDIP01117261 JAL86453.1 GAMC01013161 GAMC01013158 GAMC01013155 JAB93394.1 KB632356 ERL93410.1 GBRD01010182 JAG55642.1 APGK01041544 KB740994 ENN75955.1 KQ435687 KOX81368.1 KK854152 PTY12249.1 GL436596 EFN71752.1 KQ414705 KOC63226.1 GBGD01002822 JAC86067.1 GL888223 EGI64449.1 GFTR01003739 JAW12687.1 GDIP01171703 JAJ51699.1 KQ981261 KYN44101.1 KQ976720 KYM76708.1 ADTU01025002 ADTU01025003 ADTU01025004 ADTU01025005 GBBI01003255 JAC15457.1 GEBQ01031852 JAT08125.1 GECL01001774 JAP04350.1 ACPB03010283 KK116673 KFM68417.1 KQ983089 KYQ47651.1 GBXI01001542 JAD12750.1 KZ288191 PBC34494.1 AJWK01033970 GDIP01146504 GDIQ01198925 JAJ76898.1 JAK52800.1 JRES01001654 KNC21113.1 GDIP01132363 JAL71351.1 GDKW01002283 JAI54312.1 NEDP02004840 OWF44258.1 GDIQ01121552 JAL30174.1 GDHF01027761 JAI24553.1 AAGJ04037077 AAGJ04037078 KK107419 EZA51070.1 JH431980 GAKP01022678 JAC36274.1 KB203357 ESO84989.1 GL732540 EFX82067.1 GDIP01127664 JAL76050.1 HACG01040330 HACG01040331 CEK87195.1 CEK87196.1 GBBM01002304 JAC33114.1 JO843946 AEO35563.1 GANO01002822 JAB57049.1 GEFH01002261 JAP66320.1 GGMR01017157 MBY29776.1 KK852601 KDR20495.1 GFXV01001858 MBW13663.1 LBMM01003488 KMQ93476.1 ABLF02031720 GFWV01022810 MAA47537.1 AFYH01136520 AFYH01136521 BDGG01000019 GAV08881.1 CVRI01000004 CRK87566.1 GFPF01005608 MAA16754.1 GEDV01006702 JAP81855.1 GACK01006113 JAA58921.1 GEFM01002858 JAP72938.1 GDIP01171704 JAJ51698.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000037510
UP000218220
+ More
UP000192223 UP000245037 UP000007266 UP000215335 UP000002358 UP000076502 UP000095301 UP000030742 UP000019118 UP000053105 UP000095300 UP000000311 UP000053825 UP000007755 UP000078541 UP000078540 UP000005205 UP000085678 UP000015103 UP000054359 UP000075809 UP000242457 UP000092461 UP000037069 UP000242188 UP000007110 UP000053097 UP000005203 UP000076420 UP000030746 UP000000305 UP000027135 UP000036403 UP000007819 UP000008672 UP000186922 UP000183832
UP000192223 UP000245037 UP000007266 UP000215335 UP000002358 UP000076502 UP000095301 UP000030742 UP000019118 UP000053105 UP000095300 UP000000311 UP000053825 UP000007755 UP000078541 UP000078540 UP000005205 UP000085678 UP000015103 UP000054359 UP000075809 UP000242457 UP000092461 UP000037069 UP000242188 UP000007110 UP000053097 UP000005203 UP000076420 UP000030746 UP000000305 UP000027135 UP000036403 UP000007819 UP000008672 UP000186922 UP000183832
Interpro
SUPFAM
SSF100950
SSF100950
Gene 3D
CDD
ProteinModelPortal
H9JWK2
A2TF14
A0A0N1INN4
A0A194Q017
A0A212FC63
S4P9E4
+ More
A0A2H1WMK7 A0A0L7LDU1 A0A2A4JCA4 A0A1W4XRF1 A0A2W1BG55 A0A2P8XFX3 A0A0C9Q5I1 D6WWZ6 A0A1B6CM29 A0A336MWD1 A0A1L8E2G0 A0A1B6H374 A0A232FBU0 K7IW11 A0A336MJQ0 A0A1Y1NEX0 A0A1L8E1T4 A0A0P4W4X3 A0A1L8E1Z4 A0A1B6KH60 A0A0P5KH27 A0A0P5BVL0 A0A0P5YXZ1 A0A154NZ42 A0A0N8DM66 A0A1L8E1M6 A0A310SHD3 A0A0P6IGJ5 A0A146LI57 A0A0A9WAB3 A0A0P5U6P0 A0A1I8N1L4 W8BW14 U4UTG0 A0A0K8SQJ7 N6U308 A0A0N0BL12 A0A1I8QA94 A0A2R7VY42 E2A481 A0A0L7QXE0 A0A069DQY8 F4WMX9 A0A224XJM6 A0A0P5CSK3 A0A195FUK7 A0A151HYV3 A0A158NSK5 A0A1S3KGS3 A0A023F1S0 A0A1B6K9M7 A0A0V0G8Q8 T1HIG0 A0A087TTH8 A0A151WIL5 A0A0A1XPE9 A0A2A3ESK3 A0A1B0GKZ1 A0A0P5E676 A0A0L0BMF2 A0A0P5T1I7 A0A0N7Z8V6 A0A210Q6A3 A0A0P5RE35 A0A0K8UDB3 W4XGH6 A0A026W4R4 T1JA20 A0A088A588 A0A2C9K2K9 A0A034V1A5 V4B8D7 E9GDE3 A0A0P5TED1 A0A1I8MXD4 A0A0B7B4I1 A0A023GGW8 G3MPZ5 U5EWT6 A0A131XJM1 A0A2S2PLM6 A0A067RLS3 A0A2H8THQ6 A0A0J7KT26 J9K0I9 A0A293MPE1 H3AHH1 A0A1D1W620 A0A1J1HM86 A0A224YS02 A0A131YTI9 L7M3L3 A0A131Y354 A0A0P5CEK3
A0A2H1WMK7 A0A0L7LDU1 A0A2A4JCA4 A0A1W4XRF1 A0A2W1BG55 A0A2P8XFX3 A0A0C9Q5I1 D6WWZ6 A0A1B6CM29 A0A336MWD1 A0A1L8E2G0 A0A1B6H374 A0A232FBU0 K7IW11 A0A336MJQ0 A0A1Y1NEX0 A0A1L8E1T4 A0A0P4W4X3 A0A1L8E1Z4 A0A1B6KH60 A0A0P5KH27 A0A0P5BVL0 A0A0P5YXZ1 A0A154NZ42 A0A0N8DM66 A0A1L8E1M6 A0A310SHD3 A0A0P6IGJ5 A0A146LI57 A0A0A9WAB3 A0A0P5U6P0 A0A1I8N1L4 W8BW14 U4UTG0 A0A0K8SQJ7 N6U308 A0A0N0BL12 A0A1I8QA94 A0A2R7VY42 E2A481 A0A0L7QXE0 A0A069DQY8 F4WMX9 A0A224XJM6 A0A0P5CSK3 A0A195FUK7 A0A151HYV3 A0A158NSK5 A0A1S3KGS3 A0A023F1S0 A0A1B6K9M7 A0A0V0G8Q8 T1HIG0 A0A087TTH8 A0A151WIL5 A0A0A1XPE9 A0A2A3ESK3 A0A1B0GKZ1 A0A0P5E676 A0A0L0BMF2 A0A0P5T1I7 A0A0N7Z8V6 A0A210Q6A3 A0A0P5RE35 A0A0K8UDB3 W4XGH6 A0A026W4R4 T1JA20 A0A088A588 A0A2C9K2K9 A0A034V1A5 V4B8D7 E9GDE3 A0A0P5TED1 A0A1I8MXD4 A0A0B7B4I1 A0A023GGW8 G3MPZ5 U5EWT6 A0A131XJM1 A0A2S2PLM6 A0A067RLS3 A0A2H8THQ6 A0A0J7KT26 J9K0I9 A0A293MPE1 H3AHH1 A0A1D1W620 A0A1J1HM86 A0A224YS02 A0A131YTI9 L7M3L3 A0A131Y354 A0A0P5CEK3
PDB
3ICO
E-value=1.74691e-17,
Score=216
Ontologies
KEGG
PATHWAY
GO
PANTHER
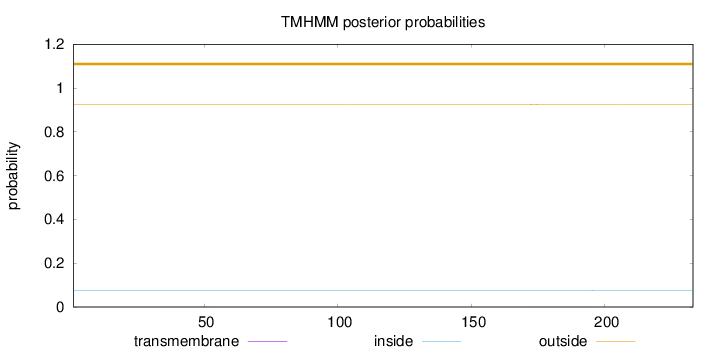
Topology
Length:
233
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02043
Exp number, first 60 AAs:
0.00492
Total prob of N-in:
0.07543
outside
1 - 233
Population Genetic Test Statistics
Pi
200.561732
Theta
207.322876
Tajima's D
-0.608052
CLR
0
CSRT
0.214089295535223
Interpretation
Uncertain
