Gene
KWMTBOMO16197 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013965
Annotation
H+_transporting_ATP_synthase_gamma_subunit_[Bombyx_mori]
Full name
ATP synthase subunit gamma
+ More
ATP synthase subunit gamma, mitochondrial
ATP synthase subunit gamma, mitochondrial
Alternative Name
F-ATPase gamma subunit
Location in the cell
Mitochondrial Reliability : 3.707
Sequence
CDS
ATGTTGGGACGCTTCGGACCGGGTGTCGGCACCCAAGTCGTGGCGGTGGTTTACCATCAGCCAAACAGGAATATGGCTACTTTGAAGGCCATTTCCATCCGTCTTAAATCTGTGAAAAATATCCAGAAAATCACACAGTCCATGAAGATGGTGTCAGCTGCTAAATACACCCGTGCTGAGCGTGACCTGAAAGCTGCTCGTCCCTATGGTGAAGGTGCAGTACAGTTCTATGAAAGGGCTGAGGTTACACCTCCCGAAGATGACCCCAAGCAATTGTTTGTTGCTATGACCTCTGACAGAGGTTTGTGCGGAGCTGTACACACTGGTGTATCCAAAGTGATCCGCAACCGTCTCAGCGAACCTGGTGCTGAGAACATCAAGGTGATCTGTGTGGGAGATAAATCTCGCGGTATCCTGCAGAGATTGTACGGAAAGCACATCATTAGTGTTGCTAATGAGATCGGACGTCTCCCACCTACTTTCTTGGACGCAAGTCAGCTGGCCACTGCTATCCTCACCTCGGGATACGAGTTTGGTTCCGGAAAGATCATTTACAACAAGTTCAAGTCTGTGGTCTCGTACGCCCAGTCCGACTTGCCCCTCTACACTAAGAAGTCTATTGAGAGCGCCAGCAAGCTGACTGCGTACGACTCTCTGGACAGCGACGTGCTCCAATCCTACACGGAGTTCTCGCTGGCCTCGCTGTTGTTCTACGCGCTGAAGGAGGGCGCCTGCTCAGAGCAGTCGTCCCGCATGACCGCCATGGACAACGCCTCCAAGAACGCCGGCGAGATGATCGACAAGCTGACCCTCACGTTCAACAGGACCCGTCAAGCTGTCATCACCAGGGAGCTCATCGAGATCATCTCTGGTGCGGCCGCCTTGGACTAA
Protein
MLGRFGPGVGTQVVAVVYHQPNRNMATLKAISIRLKSVKNIQKITQSMKMVSAAKYTRAERDLKAARPYGEGAVQFYERAEVTPPEDDPKQLFVAMTSDRGLCGAVHTGVSKVIRNRLSEPGAENIKVICVGDKSRGILQRLYGKHIISVANEIGRLPPTFLDASQLATAILTSGYEFGSGKIIYNKFKSVVSYAQSDLPLYTKKSIESASKLTAYDSLDSDVLQSYTEFSLASLLFYALKEGACSEQSSRMTAMDNASKNAGEMIDKLTLTFNRTRQAVITRELIEIISGAAALD
Summary
Subunit
F-type ATPases have 2 components, CF(1) - the catalytic core - and CF(0) - the membrane proton channel. CF(1) has five subunits: alpha(3), beta(3), gamma(1), delta(1), epsilon(1). CF(0) has three main subunits: a, b and c.
Similarity
Belongs to the ATPase gamma chain family.
Keywords
ATP synthesis
Complete proteome
Hydrogen ion transport
Ion transport
Membrane
Mitochondrion
Mitochondrion inner membrane
Reference proteome
Transit peptide
Transport
Feature
chain ATP synthase subunit gamma
Uniprot
Q1HPX4
S4NNE5
I4DJ96
A0A212F7F8
A0A194QVY4
A0A2H1WBL5
+ More
A0A3S2TJQ6 A0A2A4JBB2 A0A2W1B7N2 A0A3G1T1C7 G9D519 A0A2J7PRF8 A0A067RHY8 A0A2P8Z4D8 B0W2W3 A0A084WHB1 A0A182QIX0 A0A182NTJ6 A0A182LYU4 A0A182W7I1 A0A182UP01 A0A182L1Z8 A0A182UFE6 A0A182HV43 A0A182PD01 A0A182XZU4 Q16XK3 A0A1W4WJB5 W5JNX9 A0A182WT48 A0A2M4BVD1 A0A182RJB1 A0A2M4A2J6 A0A182F302 A0A182JWN3 Q7Q3N8 A0A2M4BWD1 A0A023ENY5 A0A2M3YZ82 T1DPU9 U5EXE4 A0A1Q3FGI6 D6WPW7 B3MTI4 A0A1Y1LVM0 A0A0M4ET73 A0A0L0C3Z3 A0A182J7L3 C6ZQR8 B4G663 Q29AN8 O01666 J3JTS0 A0A232EVT9 W8BR16 U5EWI7 K7J3Z6 V5I9Q2 B4R084 B4PPK7 B4HZE2 B3P5D7 A0A3B0JKR2 A0A0K8VMQ3 A0A034WFC0 A0A0C9PX20 A0A1W4UQ62 A0A026WWE4 E0VFT5 A0A0A1WI89 T1PF90 A0A1L8E0N4 A0A1B6BXB8 A0A1L8EEQ6 A0A1I8QC49 B4NBL2 A0A1D1ZG56 A0A0K8U3G5 B4M5T2 A0A151WJ89 E9IJZ3 T1HN70 A0A224XPP2 A0A0V0G371 A0A0P4VV79 R4FNB9 A0A069DRH5 Q1W291 A0A2R7WX91 Q6PPH6 A0A158NT33 R4WIF5 A0A195EFE8 B4JYM7 A0A195BHB1 F4WG50 A0A154PEW2 A0A0J7KT88 A0A1S4ECU0 A0A1A9VLY0
A0A3S2TJQ6 A0A2A4JBB2 A0A2W1B7N2 A0A3G1T1C7 G9D519 A0A2J7PRF8 A0A067RHY8 A0A2P8Z4D8 B0W2W3 A0A084WHB1 A0A182QIX0 A0A182NTJ6 A0A182LYU4 A0A182W7I1 A0A182UP01 A0A182L1Z8 A0A182UFE6 A0A182HV43 A0A182PD01 A0A182XZU4 Q16XK3 A0A1W4WJB5 W5JNX9 A0A182WT48 A0A2M4BVD1 A0A182RJB1 A0A2M4A2J6 A0A182F302 A0A182JWN3 Q7Q3N8 A0A2M4BWD1 A0A023ENY5 A0A2M3YZ82 T1DPU9 U5EXE4 A0A1Q3FGI6 D6WPW7 B3MTI4 A0A1Y1LVM0 A0A0M4ET73 A0A0L0C3Z3 A0A182J7L3 C6ZQR8 B4G663 Q29AN8 O01666 J3JTS0 A0A232EVT9 W8BR16 U5EWI7 K7J3Z6 V5I9Q2 B4R084 B4PPK7 B4HZE2 B3P5D7 A0A3B0JKR2 A0A0K8VMQ3 A0A034WFC0 A0A0C9PX20 A0A1W4UQ62 A0A026WWE4 E0VFT5 A0A0A1WI89 T1PF90 A0A1L8E0N4 A0A1B6BXB8 A0A1L8EEQ6 A0A1I8QC49 B4NBL2 A0A1D1ZG56 A0A0K8U3G5 B4M5T2 A0A151WJ89 E9IJZ3 T1HN70 A0A224XPP2 A0A0V0G371 A0A0P4VV79 R4FNB9 A0A069DRH5 Q1W291 A0A2R7WX91 Q6PPH6 A0A158NT33 R4WIF5 A0A195EFE8 B4JYM7 A0A195BHB1 F4WG50 A0A154PEW2 A0A0J7KT88 A0A1S4ECU0 A0A1A9VLY0
Pubmed
19121390
23622113
22651552
26354079
22118469
28756777
+ More
21967427 24845553 29403074 24438588 20966253 25244985 17510324 20920257 23761445 12364791 14747013 17210077 24945155 26483478 18362917 19820115 17994087 28004739 26108605 20496585 15632085 23185243 10731132 12537572 12537569 10071211 22516182 23537049 28648823 24495485 20075255 17550304 25348373 24508170 30249741 20566863 25830018 25315136 21282665 27129103 26334808 21347285 23691247 21719571
21967427 24845553 29403074 24438588 20966253 25244985 17510324 20920257 23761445 12364791 14747013 17210077 24945155 26483478 18362917 19820115 17994087 28004739 26108605 20496585 15632085 23185243 10731132 12537572 12537569 10071211 22516182 23537049 28648823 24495485 20075255 17550304 25348373 24508170 30249741 20566863 25830018 25315136 21282665 27129103 26334808 21347285 23691247 21719571
EMBL
BABH01018421
DQ443278
ABF51367.1
GAIX01012289
JAA80271.1
AK401364
+ More
KQ459583 BAM17986.1 KPI98617.1 AGBW02009877 OWR49666.1 KQ461103 KPJ09474.1 ODYU01007571 SOQ50453.1 RSAL01000091 RVE47997.1 NWSH01002112 PCG69139.1 KZ150244 PZC71889.1 MG846921 AXY94773.1 JF777422 AEU11767.1 NEVH01022362 PNF18917.1 KK852622 KDR19994.1 PYGN01000201 PSN51362.1 DS231829 EDS30322.1 ATLV01023800 KE525346 KFB49605.1 AXCN02000699 AXCM01002571 APCN01002493 CH477537 EAT39355.1 ADMH02000571 ETN65841.1 GGFJ01007886 MBW57027.1 GGFK01001696 MBW35017.1 AAAB01008964 EAA12854.3 GGFJ01007887 MBW57028.1 JXUM01116988 GAPW01002938 KQ566040 JAC10660.1 KXJ70434.1 GGFM01000831 MBW21582.1 GAMD01003014 JAA98576.1 GANO01002578 JAB57293.1 GFDL01008377 JAV26668.1 KQ971354 EFA07448.1 CH902623 EDV30574.1 GEZM01050230 GEZM01050229 JAV75666.1 CP012526 ALC48060.1 JRES01001003 KNC26199.1 EZ114869 ACU30922.1 CH479179 EDW23816.1 CM000070 EAL27311.1 AE014297 AY113454 Y12701 APGK01025935 BT126629 KB740605 KB632095 AEE61592.1 ENN80061.1 ERL88735.1 NNAY01001960 OXU22447.1 GAMC01010884 JAB95671.1 GANO01002937 JAB56934.1 AAZX01000987 GALX01002540 JAB65926.1 CM000364 EDX14890.1 CM000160 EDW98257.1 CH480819 EDW53399.1 CH954182 EDV53187.1 OUUW01000007 SPP82914.1 GDHF01012143 JAI40171.1 GAKP01006469 JAC52483.1 GBYB01006053 JAG75820.1 KK107085 QOIP01000008 EZA60056.1 RLU19527.1 DS235124 EEB12241.1 GBXI01015931 JAC98360.1 KA647354 AFP61983.1 GFDF01001781 JAV12303.1 GEDC01031639 JAS05659.1 GFDG01001618 JAV17181.1 CH964232 EDW81176.1 GDJX01002097 JAT65839.1 GDHF01031075 JAI21239.1 CH940652 EDW59008.1 KQ983039 KYQ47922.1 GL763908 EFZ19110.1 ACPB03017427 GFTR01004628 JAW11798.1 GECL01003595 JAP02529.1 GDKW01000647 JAI55948.1 GAHY01001359 JAA76151.1 GBGD01002379 JAC86510.1 DQ445536 ABD98774.1 KK855861 PTY24152.1 AY588070 AAT01082.1 ADTU01025481 AK417185 BAN20400.1 KQ978957 KYN27000.1 CH916377 EDV90789.1 KQ976488 KYM83613.1 GL888128 EGI66817.1 KQ434889 KZC10395.1 LBMM01003366 KMQ93607.1
KQ459583 BAM17986.1 KPI98617.1 AGBW02009877 OWR49666.1 KQ461103 KPJ09474.1 ODYU01007571 SOQ50453.1 RSAL01000091 RVE47997.1 NWSH01002112 PCG69139.1 KZ150244 PZC71889.1 MG846921 AXY94773.1 JF777422 AEU11767.1 NEVH01022362 PNF18917.1 KK852622 KDR19994.1 PYGN01000201 PSN51362.1 DS231829 EDS30322.1 ATLV01023800 KE525346 KFB49605.1 AXCN02000699 AXCM01002571 APCN01002493 CH477537 EAT39355.1 ADMH02000571 ETN65841.1 GGFJ01007886 MBW57027.1 GGFK01001696 MBW35017.1 AAAB01008964 EAA12854.3 GGFJ01007887 MBW57028.1 JXUM01116988 GAPW01002938 KQ566040 JAC10660.1 KXJ70434.1 GGFM01000831 MBW21582.1 GAMD01003014 JAA98576.1 GANO01002578 JAB57293.1 GFDL01008377 JAV26668.1 KQ971354 EFA07448.1 CH902623 EDV30574.1 GEZM01050230 GEZM01050229 JAV75666.1 CP012526 ALC48060.1 JRES01001003 KNC26199.1 EZ114869 ACU30922.1 CH479179 EDW23816.1 CM000070 EAL27311.1 AE014297 AY113454 Y12701 APGK01025935 BT126629 KB740605 KB632095 AEE61592.1 ENN80061.1 ERL88735.1 NNAY01001960 OXU22447.1 GAMC01010884 JAB95671.1 GANO01002937 JAB56934.1 AAZX01000987 GALX01002540 JAB65926.1 CM000364 EDX14890.1 CM000160 EDW98257.1 CH480819 EDW53399.1 CH954182 EDV53187.1 OUUW01000007 SPP82914.1 GDHF01012143 JAI40171.1 GAKP01006469 JAC52483.1 GBYB01006053 JAG75820.1 KK107085 QOIP01000008 EZA60056.1 RLU19527.1 DS235124 EEB12241.1 GBXI01015931 JAC98360.1 KA647354 AFP61983.1 GFDF01001781 JAV12303.1 GEDC01031639 JAS05659.1 GFDG01001618 JAV17181.1 CH964232 EDW81176.1 GDJX01002097 JAT65839.1 GDHF01031075 JAI21239.1 CH940652 EDW59008.1 KQ983039 KYQ47922.1 GL763908 EFZ19110.1 ACPB03017427 GFTR01004628 JAW11798.1 GECL01003595 JAP02529.1 GDKW01000647 JAI55948.1 GAHY01001359 JAA76151.1 GBGD01002379 JAC86510.1 DQ445536 ABD98774.1 KK855861 PTY24152.1 AY588070 AAT01082.1 ADTU01025481 AK417185 BAN20400.1 KQ978957 KYN27000.1 CH916377 EDV90789.1 KQ976488 KYM83613.1 GL888128 EGI66817.1 KQ434889 KZC10395.1 LBMM01003366 KMQ93607.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000283053
UP000218220
+ More
UP000235965 UP000027135 UP000245037 UP000002320 UP000030765 UP000075886 UP000075884 UP000075883 UP000075920 UP000075903 UP000075882 UP000075902 UP000075840 UP000075885 UP000076408 UP000008820 UP000192223 UP000000673 UP000076407 UP000075900 UP000069272 UP000075881 UP000007062 UP000069940 UP000249989 UP000007266 UP000007801 UP000092553 UP000037069 UP000075880 UP000008744 UP000001819 UP000000803 UP000019118 UP000030742 UP000215335 UP000002358 UP000000304 UP000002282 UP000001292 UP000008711 UP000268350 UP000192221 UP000053097 UP000279307 UP000009046 UP000095301 UP000095300 UP000007798 UP000008792 UP000075809 UP000015103 UP000005205 UP000078492 UP000001070 UP000078540 UP000007755 UP000076502 UP000036403 UP000079169 UP000078200
UP000235965 UP000027135 UP000245037 UP000002320 UP000030765 UP000075886 UP000075884 UP000075883 UP000075920 UP000075903 UP000075882 UP000075902 UP000075840 UP000075885 UP000076408 UP000008820 UP000192223 UP000000673 UP000076407 UP000075900 UP000069272 UP000075881 UP000007062 UP000069940 UP000249989 UP000007266 UP000007801 UP000092553 UP000037069 UP000075880 UP000008744 UP000001819 UP000000803 UP000019118 UP000030742 UP000215335 UP000002358 UP000000304 UP000002282 UP000001292 UP000008711 UP000268350 UP000192221 UP000053097 UP000279307 UP000009046 UP000095301 UP000095300 UP000007798 UP000008792 UP000075809 UP000015103 UP000005205 UP000078492 UP000001070 UP000078540 UP000007755 UP000076502 UP000036403 UP000079169 UP000078200
Interpro
Gene 3D
ProteinModelPortal
Q1HPX4
S4NNE5
I4DJ96
A0A212F7F8
A0A194QVY4
A0A2H1WBL5
+ More
A0A3S2TJQ6 A0A2A4JBB2 A0A2W1B7N2 A0A3G1T1C7 G9D519 A0A2J7PRF8 A0A067RHY8 A0A2P8Z4D8 B0W2W3 A0A084WHB1 A0A182QIX0 A0A182NTJ6 A0A182LYU4 A0A182W7I1 A0A182UP01 A0A182L1Z8 A0A182UFE6 A0A182HV43 A0A182PD01 A0A182XZU4 Q16XK3 A0A1W4WJB5 W5JNX9 A0A182WT48 A0A2M4BVD1 A0A182RJB1 A0A2M4A2J6 A0A182F302 A0A182JWN3 Q7Q3N8 A0A2M4BWD1 A0A023ENY5 A0A2M3YZ82 T1DPU9 U5EXE4 A0A1Q3FGI6 D6WPW7 B3MTI4 A0A1Y1LVM0 A0A0M4ET73 A0A0L0C3Z3 A0A182J7L3 C6ZQR8 B4G663 Q29AN8 O01666 J3JTS0 A0A232EVT9 W8BR16 U5EWI7 K7J3Z6 V5I9Q2 B4R084 B4PPK7 B4HZE2 B3P5D7 A0A3B0JKR2 A0A0K8VMQ3 A0A034WFC0 A0A0C9PX20 A0A1W4UQ62 A0A026WWE4 E0VFT5 A0A0A1WI89 T1PF90 A0A1L8E0N4 A0A1B6BXB8 A0A1L8EEQ6 A0A1I8QC49 B4NBL2 A0A1D1ZG56 A0A0K8U3G5 B4M5T2 A0A151WJ89 E9IJZ3 T1HN70 A0A224XPP2 A0A0V0G371 A0A0P4VV79 R4FNB9 A0A069DRH5 Q1W291 A0A2R7WX91 Q6PPH6 A0A158NT33 R4WIF5 A0A195EFE8 B4JYM7 A0A195BHB1 F4WG50 A0A154PEW2 A0A0J7KT88 A0A1S4ECU0 A0A1A9VLY0
A0A3S2TJQ6 A0A2A4JBB2 A0A2W1B7N2 A0A3G1T1C7 G9D519 A0A2J7PRF8 A0A067RHY8 A0A2P8Z4D8 B0W2W3 A0A084WHB1 A0A182QIX0 A0A182NTJ6 A0A182LYU4 A0A182W7I1 A0A182UP01 A0A182L1Z8 A0A182UFE6 A0A182HV43 A0A182PD01 A0A182XZU4 Q16XK3 A0A1W4WJB5 W5JNX9 A0A182WT48 A0A2M4BVD1 A0A182RJB1 A0A2M4A2J6 A0A182F302 A0A182JWN3 Q7Q3N8 A0A2M4BWD1 A0A023ENY5 A0A2M3YZ82 T1DPU9 U5EXE4 A0A1Q3FGI6 D6WPW7 B3MTI4 A0A1Y1LVM0 A0A0M4ET73 A0A0L0C3Z3 A0A182J7L3 C6ZQR8 B4G663 Q29AN8 O01666 J3JTS0 A0A232EVT9 W8BR16 U5EWI7 K7J3Z6 V5I9Q2 B4R084 B4PPK7 B4HZE2 B3P5D7 A0A3B0JKR2 A0A0K8VMQ3 A0A034WFC0 A0A0C9PX20 A0A1W4UQ62 A0A026WWE4 E0VFT5 A0A0A1WI89 T1PF90 A0A1L8E0N4 A0A1B6BXB8 A0A1L8EEQ6 A0A1I8QC49 B4NBL2 A0A1D1ZG56 A0A0K8U3G5 B4M5T2 A0A151WJ89 E9IJZ3 T1HN70 A0A224XPP2 A0A0V0G371 A0A0P4VV79 R4FNB9 A0A069DRH5 Q1W291 A0A2R7WX91 Q6PPH6 A0A158NT33 R4WIF5 A0A195EFE8 B4JYM7 A0A195BHB1 F4WG50 A0A154PEW2 A0A0J7KT88 A0A1S4ECU0 A0A1A9VLY0
PDB
2W6J
E-value=2.9351e-86,
Score=810
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Mitochondrion
Mitochondrion inner membrane
Mitochondrion inner membrane
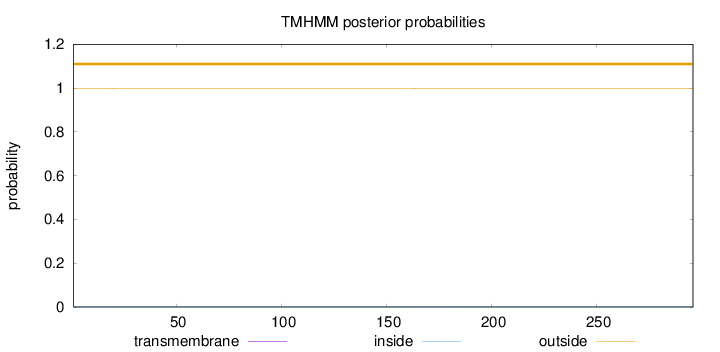
Length:
296
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02179
Exp number, first 60 AAs:
0.0066
Total prob of N-in:
0.00296
outside
1 - 296
Population Genetic Test Statistics
Pi
166.748796
Theta
135.371392
Tajima's D
0.877914
CLR
0.577228
CSRT
0.627418629068547
Interpretation
Uncertain
