Gene
KWMTBOMO16196 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013964
Annotation
vacuolar_ATPase_subunit_C_[Bombyx_mori]
Full name
V-type proton ATPase subunit C
Alternative Name
Vacuolar proton pump subunit C
Location in the cell
Cytoplasmic Reliability : 1.992
Sequence
CDS
ATGACTGAATACTGGGTGATCAGTGCCCCTGGCGACAAGACCTGTCAGCAGACTTGGGATACCCTGAACAATGCCACCAAATCAGGCAACCTCAGTGTCAACTATAAATTCCCTATTCCTGACCTTAAGGTGGGAACTTTGGATCAGTTGGTGGGGCTGTCAGATGATCTGGGCAAGCTTGACACTTTCGTCGAAGGTGTGACCAGGAAGGTAGCCCAGTACCTTGGAGAGGTACTTGAAGATCAGCGCGATAAGCTCCATGAAAACTTGATGGCCAATAACAGTGACCTCCCCACTTATTTGACTCGCTTCCAATGGGACATGGCTAAGTACCCCATAAAGCAGAGTCTGCGGAATATCGCCGACATCATAAGCAAACAGGTCGGACAGATCGACGCGGATCTGAAGGTCAAGTCCTCCGCCTACAACGCTCTCAAAGGAAACCTACACAATTTAGAGAAGAAACAGACCGGGAGCTTGTTGACCCGCAACTTGGCCGATCTAGTCAAGAAGGAGCACTTCATCTTGGACAGCGAGTACCTGACCACACTCCTGGTCATCGTGCCCAAGTCAATGTTCAACGACTGGAATGCCAACTACGAGAAGATAACGGACATGATCGTGCCGCGCTCCACCCAGCTCGTCCACCAGGACAACGACTACGGACTCTTCACCGTCACCCTCTTCAAGAAGGTTGCGGACGAGTTCAAGCTGCACGCTCGCGAGCGCAAGTTCGTCGTGCGCGAGTTCGCTTACAACGAGGCCGACCTGCTCGCCGGCAAGAACGAGATCACCAAGCTCGTCACCGACAAGAAGAAGCAGTTTGGACCGCTGGTGCGATGGTTGAAGGTGAACTTCTCCGAGTGCTTCTGCGCTTGGATCCACGTTAAAGCTCTGCGTGTGTTCGTCGAATCCGTTTTGAGATACGGTCTGCCGGTGAACTTCCAGGCCGTGGTGATGGTCCCGGCGCGCAAGAGCATGAAGAAGCTGCGCGACCTCCTCAACCAGCTCTACGCGCACCTCGACCACTCCGCGCACGCGCACTCCGCCGCCGCGCCGGACAGCGTGGAACTAGCCGGCCTAGGTTTCGGTCAATCGGAATATTTCCCGTATGTTTTCTATAAGATCAACATCGACATGATCGAGAAATCGTCGGCCTAA
Protein
MTEYWVISAPGDKTCQQTWDTLNNATKSGNLSVNYKFPIPDLKVGTLDQLVGLSDDLGKLDTFVEGVTRKVAQYLGEVLEDQRDKLHENLMANNSDLPTYLTRFQWDMAKYPIKQSLRNIADIISKQVGQIDADLKVKSSAYNALKGNLHNLEKKQTGSLLTRNLADLVKKEHFILDSEYLTTLLVIVPKSMFNDWNANYEKITDMIVPRSTQLVHQDNDYGLFTVTLFKKVADEFKLHARERKFVVREFAYNEADLLAGKNEITKLVTDKKKQFGPLVRWLKVNFSECFCAWIHVKALRVFVESVLRYGLPVNFQAVVMVPARKSMKKLRDLLNQLYAHLDHSAHAHSAAAPDSVELAGLGFGQSEYFPYVFYKINIDMIEKSSA
Summary
Description
Subunit of the peripheral V1 complex of vacuolar ATPase. Subunit C is necessary for the assembly of the catalytic sector of the enzyme and is likely to have a specific function in its catalytic activity. V-ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells.
Subunit of the peripheral V1 complex of vacuolar ATPase. Subunit C is necessary for the assembly of the catalytic sector of the enzyme and is likely to have a specific function in its catalytic activity. V-ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells (By similarity).
Subunit of the peripheral V1 complex of vacuolar ATPase. Subunit C is necessary for the assembly of the catalytic sector of the enzyme and is likely to have a specific function in its catalytic activity. V-ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells (By similarity).
Subunit
V-ATPase is a heteromultimeric enzyme composed of a peripheral catalytic V1 complex (components A to H) attached to an integral membrane V0 proton pore complex.
V-ATPase is a heteromultimeric enzyme composed of a peripheral catalytic V1 complex (components A to H) attached to an integral membrane V0 proton pore complex (components: a, c, c', c'' and d).
V-ATPase is a heteromultimeric enzyme composed of a peripheral catalytic V1 complex (components A to H) attached to an integral membrane V0 proton pore complex (components: a, c, c', c'' and d).
Similarity
Belongs to the V-ATPase C subunit family.
Keywords
Hydrogen ion transport
Ion transport
Transport
Alternative splicing
Complete proteome
Reference proteome
Feature
chain V-type proton ATPase subunit C
splice variant In isoform A.
splice variant In isoform A.
Uniprot
Q2F673
I4DR85
S4PV58
A0A076PXI2
Q9U5N1
A0A2W1B9M2
+ More
A0A2R8G1V8 D6WTD4 A0A2A4J9R2 A0A2P8XH01 A0A2J7RHD8 A0A1B6EYU4 A4KYS3 A0A232EJE5 A0A067QZJ7 A0A0C9RWM2 A0A026WRR5 A0A1L8DF84 F4X7W1 V5I8V6 E2BTC0 B4P685 Q9V7N5-2 A0A1Y1MCX9 B4HSW7 B4QHX5 A0A0Q9WA32 A0A1Q3FY65 J9K1C1 A0A1I9WLJ0 A0A0Q9VXY6 U4U920 A0A1J1HYK5 A0A2Z4EF26 A0A2S2NGE2 A0A1L8ECS3 A0A1W4UXN1 A0A3B0JDV2 B4GAC0 A0A182H7D6 B3MFN3 A0A1W4WC36 A0A034WRR8 V9IAU4 T1DI77 U5EWK4 A0A1L8EHF0 W8BXQ6 A0A1L8EH24 A0A1B6CKL6 A0A0A1WNQ5 A0A182WLA5 A0A182M2Q2 A0A182RZ07 T1PP69 A0A1I8QEB7 E1ZUV1 A0A0K8TRG7 A0A084WE88 A0A182UXR0 A0A182TV74 A0A182XMD6 A0A182KPZ6 Q7Q6H4 A0A182I270 A0A182YJ84 A0A0V0GCQ3 A0A182JMH7 A0A182P2V7 A0A2M4BR66 A0A069DTM9 A0A0P4VUX5 A0A2M4A4V6 A0A2M4CTD0 A0A336M2X5 A0A336LZ40 A0A2M3Z182 T1DH19 A0A1S4ELA8 D3TMG7 A0A1A9XWH0 A0A0A9W801 A0A2J7RHE6 A0A1A9VU46 A0A0K8SJZ5 A0A0K8SE36 A0A146MHI9 A0A0A9YAE1 Q1HQL5 A0A1L8EAD2 A0A224XSB6 A0A0P5VRC2 E9FQR8 A0A0N8BEA6 A0A164PT35 A0A0P6I9H9 A0A0P5XTA3 A0A0P5J7Y9 A0A0P6BBZ7 H9JWQ0
A0A2R8G1V8 D6WTD4 A0A2A4J9R2 A0A2P8XH01 A0A2J7RHD8 A0A1B6EYU4 A4KYS3 A0A232EJE5 A0A067QZJ7 A0A0C9RWM2 A0A026WRR5 A0A1L8DF84 F4X7W1 V5I8V6 E2BTC0 B4P685 Q9V7N5-2 A0A1Y1MCX9 B4HSW7 B4QHX5 A0A0Q9WA32 A0A1Q3FY65 J9K1C1 A0A1I9WLJ0 A0A0Q9VXY6 U4U920 A0A1J1HYK5 A0A2Z4EF26 A0A2S2NGE2 A0A1L8ECS3 A0A1W4UXN1 A0A3B0JDV2 B4GAC0 A0A182H7D6 B3MFN3 A0A1W4WC36 A0A034WRR8 V9IAU4 T1DI77 U5EWK4 A0A1L8EHF0 W8BXQ6 A0A1L8EH24 A0A1B6CKL6 A0A0A1WNQ5 A0A182WLA5 A0A182M2Q2 A0A182RZ07 T1PP69 A0A1I8QEB7 E1ZUV1 A0A0K8TRG7 A0A084WE88 A0A182UXR0 A0A182TV74 A0A182XMD6 A0A182KPZ6 Q7Q6H4 A0A182I270 A0A182YJ84 A0A0V0GCQ3 A0A182JMH7 A0A182P2V7 A0A2M4BR66 A0A069DTM9 A0A0P4VUX5 A0A2M4A4V6 A0A2M4CTD0 A0A336M2X5 A0A336LZ40 A0A2M3Z182 T1DH19 A0A1S4ELA8 D3TMG7 A0A1A9XWH0 A0A0A9W801 A0A2J7RHE6 A0A1A9VU46 A0A0K8SJZ5 A0A0K8SE36 A0A146MHI9 A0A0A9YAE1 Q1HQL5 A0A1L8EAD2 A0A224XSB6 A0A0P5VRC2 E9FQR8 A0A0N8BEA6 A0A164PT35 A0A0P6I9H9 A0A0P5XTA3 A0A0P5J7Y9 A0A0P6BBZ7 H9JWQ0
Pubmed
22651552
23622113
11030595
28756777
18362917
19820115
+ More
29403074 28648823 24845553 24508170 30249741 21719571 20798317 17994087 17550304 9584098 10731132 12537572 12537569 28004739 22936249 27538518 23537049 26483478 25348373 24330624 24495485 25830018 25315136 26369729 24438588 20966253 12364791 14747013 17210077 25244985 26334808 27129103 20353571 25401762 26823975 17204158 21292972 19121390
29403074 28648823 24845553 24508170 30249741 21719571 20798317 17994087 17550304 9584098 10731132 12537572 12537569 28004739 22936249 27538518 23537049 26483478 25348373 24330624 24495485 25830018 25315136 26369729 24438588 20966253 12364791 14747013 17210077 25244985 26334808 27129103 20353571 25401762 26823975 17204158 21292972 19121390
EMBL
DQ311199
ABD36144.1
AK404992
BAM20425.1
GAIX01009113
JAA83447.1
+ More
KJ854407 AIJ50380.1 AJ249388 KZ150244 PZC71888.1 KY921846 AVP73918.1 KQ971352 EFA06700.2 NWSH01002385 PCG68418.1 PYGN01002161 PSN31288.1 NEVH01003743 PNF40255.1 GECZ01026617 JAS43152.1 EF156436 ABO61291.1 NNAY01004023 OXU18483.1 KK853153 KDR10550.1 GBYB01013434 GBYB01013435 JAG83201.1 JAG83202.1 KK107128 QOIP01000006 EZA58381.1 RLU21282.1 GFDF01008978 JAV05106.1 GL888900 EGI57447.1 GALX01004020 JAB64446.1 GL450389 EFN81041.1 CM000158 EDW91935.2 AF006655 AE013599 AY061038 AY102676 BT015974 GEZM01039785 JAV81197.1 CH480816 EDW48131.1 CM000362 CM002911 EDX07346.1 KMY94245.1 CH940667 KRF77675.1 GFDL01002544 JAV32501.1 ABLF02008149 KU932374 APA34010.1 KRF77676.1 KB631912 ERL87116.1 CVRI01000020 CRK91209.1 MF537670 AWV49185.1 GGMR01003383 MBY16002.1 GFDG01002278 JAV16521.1 OUUW01000001 SPP73430.1 CH479181 EDW31872.1 JXUM01028212 JXUM01028213 JXUM01028214 KQ560814 KXJ80786.1 CH902619 EDV37723.1 GAKP01002489 JAC56463.1 JR037434 JR037435 AEY57767.1 AEY57768.1 GALA01001091 JAA93761.1 GANO01001376 JAB58495.1 GFDG01000776 JAV18023.1 GAMC01012551 JAB94004.1 GFDG01000777 JAV18022.1 GEDC01024175 GEDC01023370 JAS13123.1 JAS13928.1 GBXI01017216 GBXI01014239 JAC97075.1 JAD00053.1 AXCM01007399 KA645139 KA649793 AFP64422.1 GL434297 EFN75043.1 GDAI01000875 JAI16728.1 ATLV01023195 KE525341 KFB48532.1 AAAB01008960 EAA11948.4 APCN01000126 GECL01000596 JAP05528.1 GGFJ01006332 MBW55473.1 GBGD01001768 JAC87121.1 GDKW01000757 JAI55838.1 GGFK01002431 MBW35752.1 GGFL01004426 MBW68604.1 UFQT01000465 SSX24592.1 UFQT01000243 SSX22281.1 GGFM01001523 MBW22274.1 GAMD01002541 JAA99049.1 EZ422619 ADD18895.1 GBHO01040058 GBRD01013622 GDHC01018586 JAG03546.1 JAG52204.1 JAQ00043.1 PNF40254.1 GBRD01012241 JAG53583.1 GBRD01014281 GBRD01014280 JAG51546.1 GDHC01000202 JAQ18427.1 GBHO01017094 GDHC01015317 GDHC01006382 JAG26510.1 JAQ03312.1 JAQ12247.1 DQ440429 ABF18462.1 GFDG01003108 JAV15691.1 GFTR01005545 JAW10881.1 GDIP01110948 JAL92766.1 GL732523 EFX90034.1 GDIQ01186211 JAK65514.1 LRGB01002568 KZS07115.1 GDIQ01024374 JAN70363.1 GDIP01067562 JAM36153.1 GDIQ01203359 JAK48366.1 GDIP01031396 JAM72319.1 BABH01018421
KJ854407 AIJ50380.1 AJ249388 KZ150244 PZC71888.1 KY921846 AVP73918.1 KQ971352 EFA06700.2 NWSH01002385 PCG68418.1 PYGN01002161 PSN31288.1 NEVH01003743 PNF40255.1 GECZ01026617 JAS43152.1 EF156436 ABO61291.1 NNAY01004023 OXU18483.1 KK853153 KDR10550.1 GBYB01013434 GBYB01013435 JAG83201.1 JAG83202.1 KK107128 QOIP01000006 EZA58381.1 RLU21282.1 GFDF01008978 JAV05106.1 GL888900 EGI57447.1 GALX01004020 JAB64446.1 GL450389 EFN81041.1 CM000158 EDW91935.2 AF006655 AE013599 AY061038 AY102676 BT015974 GEZM01039785 JAV81197.1 CH480816 EDW48131.1 CM000362 CM002911 EDX07346.1 KMY94245.1 CH940667 KRF77675.1 GFDL01002544 JAV32501.1 ABLF02008149 KU932374 APA34010.1 KRF77676.1 KB631912 ERL87116.1 CVRI01000020 CRK91209.1 MF537670 AWV49185.1 GGMR01003383 MBY16002.1 GFDG01002278 JAV16521.1 OUUW01000001 SPP73430.1 CH479181 EDW31872.1 JXUM01028212 JXUM01028213 JXUM01028214 KQ560814 KXJ80786.1 CH902619 EDV37723.1 GAKP01002489 JAC56463.1 JR037434 JR037435 AEY57767.1 AEY57768.1 GALA01001091 JAA93761.1 GANO01001376 JAB58495.1 GFDG01000776 JAV18023.1 GAMC01012551 JAB94004.1 GFDG01000777 JAV18022.1 GEDC01024175 GEDC01023370 JAS13123.1 JAS13928.1 GBXI01017216 GBXI01014239 JAC97075.1 JAD00053.1 AXCM01007399 KA645139 KA649793 AFP64422.1 GL434297 EFN75043.1 GDAI01000875 JAI16728.1 ATLV01023195 KE525341 KFB48532.1 AAAB01008960 EAA11948.4 APCN01000126 GECL01000596 JAP05528.1 GGFJ01006332 MBW55473.1 GBGD01001768 JAC87121.1 GDKW01000757 JAI55838.1 GGFK01002431 MBW35752.1 GGFL01004426 MBW68604.1 UFQT01000465 SSX24592.1 UFQT01000243 SSX22281.1 GGFM01001523 MBW22274.1 GAMD01002541 JAA99049.1 EZ422619 ADD18895.1 GBHO01040058 GBRD01013622 GDHC01018586 JAG03546.1 JAG52204.1 JAQ00043.1 PNF40254.1 GBRD01012241 JAG53583.1 GBRD01014281 GBRD01014280 JAG51546.1 GDHC01000202 JAQ18427.1 GBHO01017094 GDHC01015317 GDHC01006382 JAG26510.1 JAQ03312.1 JAQ12247.1 DQ440429 ABF18462.1 GFDG01003108 JAV15691.1 GFTR01005545 JAW10881.1 GDIP01110948 JAL92766.1 GL732523 EFX90034.1 GDIQ01186211 JAK65514.1 LRGB01002568 KZS07115.1 GDIQ01024374 JAN70363.1 GDIP01067562 JAM36153.1 GDIQ01203359 JAK48366.1 GDIP01031396 JAM72319.1 BABH01018421
Proteomes
UP000007266
UP000218220
UP000245037
UP000235965
UP000215335
UP000027135
+ More
UP000053097 UP000279307 UP000007755 UP000008237 UP000002282 UP000000803 UP000001292 UP000000304 UP000008792 UP000007819 UP000030742 UP000183832 UP000192221 UP000268350 UP000008744 UP000069940 UP000249989 UP000007801 UP000192223 UP000075920 UP000075883 UP000075900 UP000095301 UP000095300 UP000000311 UP000030765 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000076408 UP000075880 UP000075885 UP000079169 UP000092443 UP000078200 UP000000305 UP000076858 UP000005204
UP000053097 UP000279307 UP000007755 UP000008237 UP000002282 UP000000803 UP000001292 UP000000304 UP000008792 UP000007819 UP000030742 UP000183832 UP000192221 UP000268350 UP000008744 UP000069940 UP000249989 UP000007801 UP000192223 UP000075920 UP000075883 UP000075900 UP000095301 UP000095300 UP000000311 UP000030765 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000076408 UP000075880 UP000075885 UP000079169 UP000092443 UP000078200 UP000000305 UP000076858 UP000005204
Pfam
PF03223 V-ATPase_C
SUPFAM
SSF118203
SSF118203
ProteinModelPortal
Q2F673
I4DR85
S4PV58
A0A076PXI2
Q9U5N1
A0A2W1B9M2
+ More
A0A2R8G1V8 D6WTD4 A0A2A4J9R2 A0A2P8XH01 A0A2J7RHD8 A0A1B6EYU4 A4KYS3 A0A232EJE5 A0A067QZJ7 A0A0C9RWM2 A0A026WRR5 A0A1L8DF84 F4X7W1 V5I8V6 E2BTC0 B4P685 Q9V7N5-2 A0A1Y1MCX9 B4HSW7 B4QHX5 A0A0Q9WA32 A0A1Q3FY65 J9K1C1 A0A1I9WLJ0 A0A0Q9VXY6 U4U920 A0A1J1HYK5 A0A2Z4EF26 A0A2S2NGE2 A0A1L8ECS3 A0A1W4UXN1 A0A3B0JDV2 B4GAC0 A0A182H7D6 B3MFN3 A0A1W4WC36 A0A034WRR8 V9IAU4 T1DI77 U5EWK4 A0A1L8EHF0 W8BXQ6 A0A1L8EH24 A0A1B6CKL6 A0A0A1WNQ5 A0A182WLA5 A0A182M2Q2 A0A182RZ07 T1PP69 A0A1I8QEB7 E1ZUV1 A0A0K8TRG7 A0A084WE88 A0A182UXR0 A0A182TV74 A0A182XMD6 A0A182KPZ6 Q7Q6H4 A0A182I270 A0A182YJ84 A0A0V0GCQ3 A0A182JMH7 A0A182P2V7 A0A2M4BR66 A0A069DTM9 A0A0P4VUX5 A0A2M4A4V6 A0A2M4CTD0 A0A336M2X5 A0A336LZ40 A0A2M3Z182 T1DH19 A0A1S4ELA8 D3TMG7 A0A1A9XWH0 A0A0A9W801 A0A2J7RHE6 A0A1A9VU46 A0A0K8SJZ5 A0A0K8SE36 A0A146MHI9 A0A0A9YAE1 Q1HQL5 A0A1L8EAD2 A0A224XSB6 A0A0P5VRC2 E9FQR8 A0A0N8BEA6 A0A164PT35 A0A0P6I9H9 A0A0P5XTA3 A0A0P5J7Y9 A0A0P6BBZ7 H9JWQ0
A0A2R8G1V8 D6WTD4 A0A2A4J9R2 A0A2P8XH01 A0A2J7RHD8 A0A1B6EYU4 A4KYS3 A0A232EJE5 A0A067QZJ7 A0A0C9RWM2 A0A026WRR5 A0A1L8DF84 F4X7W1 V5I8V6 E2BTC0 B4P685 Q9V7N5-2 A0A1Y1MCX9 B4HSW7 B4QHX5 A0A0Q9WA32 A0A1Q3FY65 J9K1C1 A0A1I9WLJ0 A0A0Q9VXY6 U4U920 A0A1J1HYK5 A0A2Z4EF26 A0A2S2NGE2 A0A1L8ECS3 A0A1W4UXN1 A0A3B0JDV2 B4GAC0 A0A182H7D6 B3MFN3 A0A1W4WC36 A0A034WRR8 V9IAU4 T1DI77 U5EWK4 A0A1L8EHF0 W8BXQ6 A0A1L8EH24 A0A1B6CKL6 A0A0A1WNQ5 A0A182WLA5 A0A182M2Q2 A0A182RZ07 T1PP69 A0A1I8QEB7 E1ZUV1 A0A0K8TRG7 A0A084WE88 A0A182UXR0 A0A182TV74 A0A182XMD6 A0A182KPZ6 Q7Q6H4 A0A182I270 A0A182YJ84 A0A0V0GCQ3 A0A182JMH7 A0A182P2V7 A0A2M4BR66 A0A069DTM9 A0A0P4VUX5 A0A2M4A4V6 A0A2M4CTD0 A0A336M2X5 A0A336LZ40 A0A2M3Z182 T1DH19 A0A1S4ELA8 D3TMG7 A0A1A9XWH0 A0A0A9W801 A0A2J7RHE6 A0A1A9VU46 A0A0K8SJZ5 A0A0K8SE36 A0A146MHI9 A0A0A9YAE1 Q1HQL5 A0A1L8EAD2 A0A224XSB6 A0A0P5VRC2 E9FQR8 A0A0N8BEA6 A0A164PT35 A0A0P6I9H9 A0A0P5XTA3 A0A0P5J7Y9 A0A0P6BBZ7 H9JWQ0
PDB
6O7X
E-value=5.64121e-58,
Score=568
Ontologies
PATHWAY
GO
PANTHER
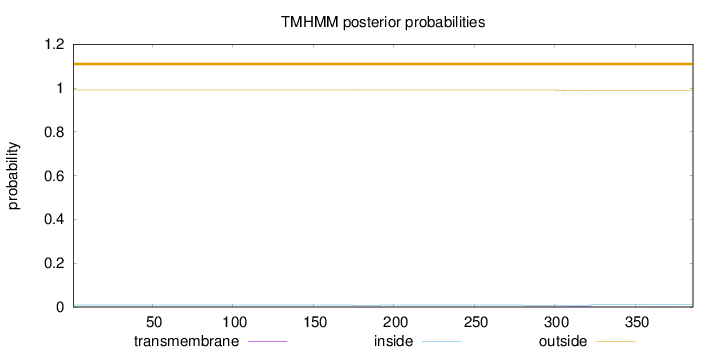
Topology
Length:
386
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06385
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00858
outside
1 - 386
Population Genetic Test Statistics
Pi
164.266289
Theta
165.960707
Tajima's D
0.032545
CLR
12.827073
CSRT
0.379931003449827
Interpretation
Uncertain
