Gene
KWMTBOMO16189
Pre Gene Modal
BGIBMGA013926
Annotation
PREDICTED:_facilitated_trehalose_transporter_Tret1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.964
Sequence
CDS
ATGGGACAATGCGGAGGCGTACCCTTACCGGGGAAAAACGTCCGTAACATCCTGGCGGCTCTTGCTGTGTCCCTTGGGCCGTTCGCAGCGGGTCTCAGTAAAGGCTACATGTCCCCAGCATTGGCGTCGATGCAGGATACAAGGATACACGATGCCTTCTCGGTTAGCGACCAACAAGCGAGTTGGATAGCTAGCCTTAGTTTATTAGGAGCCTTTATCGGTGGCATTATGGGCGGGATGGTGATGCGGTTTGGCAGAAGGACGGTGTTGCTAGCTACCGCCCTACCCTACACACTGGCGTGGCTGGCCACGGCGCTATCAAGCAGCGTGCACATGGTATCAGCCACCTCGTTCTTCGGGGGAATGCTGATCTGCTGTATAAATATGACGACACAGGTATACGTCACAGAAATAGCAGTTCCCGAGATCAGAGGATGCCTCAGCTCCGTATTGAAGATCCTCAGTCAGATCGGCATCCTAATATCATTCACTTTGGGAGCGTACCTCAACTGGTACCAACTCGCTCTGATGGTAGCCGCCGCCCCGATACTCTTATTCTTTGCTCTCTTCTTCATACCAGAAACTCCTTCTTCATTGCTCCTGAGAGGAAGGGAAACTGAAGCTGAGGATAGTCTGCAGTGGCTCAGGGGACCAGACGCGGACGTACGCCAAGAACTAGCCACAATCAGGACCAACATATTAGCCAGTAAACTGTCAAATGGGACGAACACGGGCAAATTCAAGGTATTGCTCTCGAAGCGGCTGACCAGACCAGTACTCATCACGTGCGGCCTAATGTTCTTCCAGAGATTCACTGGAGCAAACCTATTCAATTTCTACGCAGTCAAAATATTCAAGAAGACCCTAAGATCTATGAACCCTCATGGAGGGGCAATAGCGACTAGCGTTGCGCAGCTGTTGGCTTCTTGTTTGTCGGGGATGCTGATCGACAACATAGGAAGACTACCGCTTCTCATGATAAGCGGGGTGATGATGTCCATAGCCTTAGCCGGCTTCGGTTCTTACGCTTACTACGAAGAAGTTATAAGAAATACGACCGATTTAACAAGCGCCGAACCAGGATCATACGACTGGATCCCCTTGCTCTGTGTCCTGACGTTTACCATAGCCTTTTCGTTAGGCATCAGTCCGATTTCGTCTCTATTGATTGGGGAGCTGTTCCCTCTGGAGTACAGGAGTACTGGTAGCGCATTAGCGACGAGTTTCAGTCATTTGTGCGGATTCGTGAATGTTAAAACCGCCGCAGACTTCCAAGATCAAATAGGGCTCTACGGTCTGTTCTGGATGTACGCAGGTATCTCGGTCCTCTGTCTGCTGTTCGTGGTGCTATTCGTTCCGGAAACGAAGGGCCGCGAAATAGACGAGATGGACCCGAAATACGTAGAATCTTTGTGCATAAATCGATAA
Protein
MGQCGGVPLPGKNVRNILAALAVSLGPFAAGLSKGYMSPALASMQDTRIHDAFSVSDQQASWIASLSLLGAFIGGIMGGMVMRFGRRTVLLATALPYTLAWLATALSSSVHMVSATSFFGGMLICCINMTTQVYVTEIAVPEIRGCLSSVLKILSQIGILISFTLGAYLNWYQLALMVAAAPILLFFALFFIPETPSSLLLRGRETEAEDSLQWLRGPDADVRQELATIRTNILASKLSNGTNTGKFKVLLSKRLTRPVLITCGLMFFQRFTGANLFNFYAVKIFKKTLRSMNPHGGAIATSVAQLLASCLSGMLIDNIGRLPLLMISGVMMSIALAGFGSYAYYEEVIRNTTDLTSAEPGSYDWIPLLCVLTFTIAFSLGISPISSLLIGELFPLEYRSTGSALATSFSHLCGFVNVKTAADFQDQIGLYGLFWMYAGISVLCLLFVVLFVPETKGREIDEMDPKYVESLCINR
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9JWL2
A0A0N1IQH6
A0A2H1VAG8
A0A2A4JI40
A0A212F3X0
A0A194Q0T7
+ More
A0A2W1B6A7 A0A067RJL1 A0A2H1X014 A0A212EVB4 A0A2A4JIX3 A0A2W1BFS2 A0A0N1PK48 A0A194PZZ5 A0A0L7KX71 A0A1B6DDZ3 A0A2J7Q9G2 A0A2P8YW86 Q176C5 A0A1J1I0H3 A0A0P6IYT0 Q7PVH7 T1IEH0 A0A0A8JAF9 A0A182KBT0 A0A1S4H1F3 A0A182VUH5 A0A182RIN3 A0A0L7R1T9 A0A1B0CER3 E2BC95 A0A088A5V8 A0A195DQY8 W5JBS0 A0A151X9N0 A0A154P451 A0A195BF50 A0A158NBQ8 A0A0M9AAU7 H9JWL4 A0A182NU56 A0A182HGG5 A0A195FD64 E1ZVD8 A0A182VIP1 A0A182X0S9 A0A182U3K1 A0A2R7VW70 A0A182L6E7 A0A182MC20 A0A182PCF2 A0A182YDP8 A0A182QFL9 E0VQR0 A0A1B0DDL8 A0A1S4H0K6 A0A0J7LAB8 A0A3L8DSZ0 A0A151IMC3 A0A182J1P4 A0A084VWG3 A0A2A3EIX5 F4X880 A0A182F4Q8 A0A026W6L6 A0A182S8S4 A0A1B6HG68 A0A0N8AV33 A0A0N7ZK58 A0A1W4WQM3 A0A0P5GJM1 A0A0N8E7Y3 A0A226EMJ4 A0A0P5I1M3 K7JNF8 E9H7T9 A0A1D2N708 A0A0K2USY8 A0A310SBY7 N6T9V7 A0A0P6HV98 U4UTZ1 A0A0P4YV72 A0A0P4VZ78 A0A139WEZ8 A0A226EJU1 A0A232F1L7 A0A1L8DE67 A0A0P5JAK1 A0A2P2HWM8 A0A310SDC8 W8ASC7 A0A154PM32 A0A2A3EQZ2 V9IDH9 K7J7U8 A0A1B0C869 A0A195CZT9
A0A2W1B6A7 A0A067RJL1 A0A2H1X014 A0A212EVB4 A0A2A4JIX3 A0A2W1BFS2 A0A0N1PK48 A0A194PZZ5 A0A0L7KX71 A0A1B6DDZ3 A0A2J7Q9G2 A0A2P8YW86 Q176C5 A0A1J1I0H3 A0A0P6IYT0 Q7PVH7 T1IEH0 A0A0A8JAF9 A0A182KBT0 A0A1S4H1F3 A0A182VUH5 A0A182RIN3 A0A0L7R1T9 A0A1B0CER3 E2BC95 A0A088A5V8 A0A195DQY8 W5JBS0 A0A151X9N0 A0A154P451 A0A195BF50 A0A158NBQ8 A0A0M9AAU7 H9JWL4 A0A182NU56 A0A182HGG5 A0A195FD64 E1ZVD8 A0A182VIP1 A0A182X0S9 A0A182U3K1 A0A2R7VW70 A0A182L6E7 A0A182MC20 A0A182PCF2 A0A182YDP8 A0A182QFL9 E0VQR0 A0A1B0DDL8 A0A1S4H0K6 A0A0J7LAB8 A0A3L8DSZ0 A0A151IMC3 A0A182J1P4 A0A084VWG3 A0A2A3EIX5 F4X880 A0A182F4Q8 A0A026W6L6 A0A182S8S4 A0A1B6HG68 A0A0N8AV33 A0A0N7ZK58 A0A1W4WQM3 A0A0P5GJM1 A0A0N8E7Y3 A0A226EMJ4 A0A0P5I1M3 K7JNF8 E9H7T9 A0A1D2N708 A0A0K2USY8 A0A310SBY7 N6T9V7 A0A0P6HV98 U4UTZ1 A0A0P4YV72 A0A0P4VZ78 A0A139WEZ8 A0A226EJU1 A0A232F1L7 A0A1L8DE67 A0A0P5JAK1 A0A2P2HWM8 A0A310SDC8 W8ASC7 A0A154PM32 A0A2A3EQZ2 V9IDH9 K7J7U8 A0A1B0C869 A0A195CZT9
Pubmed
EMBL
BABH01018390
KQ459630
KPJ20529.1
ODYU01001498
SOQ37791.1
NWSH01001318
+ More
PCG71727.1 AGBW02010477 OWR48436.1 KQ459583 KPI98609.1 KZ150284 PZC71619.1 KK852471 KDR23173.1 ODYU01012334 SOQ58592.1 AGBW02012232 OWR45394.1 PCG71726.1 PZC71620.1 KPJ20528.1 KPI98608.1 JTDY01004693 KOB67868.1 GEDC01013448 GEDC01009429 JAS23850.1 JAS27869.1 NEVH01016358 PNF25221.1 PYGN01000324 PSN48516.1 CH477390 EAT41976.1 CVRI01000038 CRK93847.1 GDUN01000912 JAN95007.1 AAAB01008984 EAA15072.4 ACPB03017604 AB901066 BAQ02366.1 KQ414667 KOC64832.1 AJWK01009124 GL447257 EFN86631.1 KQ980581 KYN15288.1 ADMH02001962 ETN60224.1 KQ982373 KYQ56988.1 KQ434809 KZC06729.1 KQ976500 KYM83206.1 ADTU01011283 KQ435697 KOX80765.1 BABH01018387 APCN01004224 KQ981673 KYN38343.1 GL434492 EFN74848.1 KK854126 PTY11753.1 AXCM01000161 AXCN02000773 DS235430 EEB15716.1 AJVK01005422 LBMM01000140 KMR04783.1 QOIP01000004 RLU23501.1 KQ977059 KYN06045.1 ATLV01017584 KE525174 KFB42307.1 KZ288229 PBC31677.1 GL888932 EGI57211.1 KK107372 EZA51700.1 GECU01034016 JAS73690.1 GDIQ01239995 JAK11730.1 GDIP01237473 GDIP01204568 GDIP01121437 GDIP01093679 GDIP01083749 GDIP01079601 LRGB01000311 JAI85928.1 KZS19803.1 GDIQ01241535 JAK10190.1 GDIQ01053004 JAN41733.1 LNIX01000003 OXA58357.1 GDIQ01225036 JAK26689.1 GL732601 EFX72237.1 LJIJ01000174 ODN01053.1 HACA01023814 CDW41175.1 KQ784025 OAD51877.1 APGK01045776 KB741039 ENN74518.1 GDIQ01014409 JAN80328.1 KB632367 ERL93666.1 GDIP01222290 JAJ01112.1 GDRN01105627 JAI57703.1 KQ971354 KYB26540.1 OXA57021.1 NNAY01001300 OXU24432.1 GFDF01009470 JAV04614.1 GDIQ01202583 JAK49142.1 IACF01000422 LAB66198.1 KQ775154 OAD52283.1 GAMC01019093 GAMC01019092 JAB87463.1 KQ434977 KZC12919.1 KZ288203 PBC33461.1 JR039436 AEY58702.1 AAZX01001793 AJWK01000224 AJWK01000225 KQ977041 KYN06178.1
PCG71727.1 AGBW02010477 OWR48436.1 KQ459583 KPI98609.1 KZ150284 PZC71619.1 KK852471 KDR23173.1 ODYU01012334 SOQ58592.1 AGBW02012232 OWR45394.1 PCG71726.1 PZC71620.1 KPJ20528.1 KPI98608.1 JTDY01004693 KOB67868.1 GEDC01013448 GEDC01009429 JAS23850.1 JAS27869.1 NEVH01016358 PNF25221.1 PYGN01000324 PSN48516.1 CH477390 EAT41976.1 CVRI01000038 CRK93847.1 GDUN01000912 JAN95007.1 AAAB01008984 EAA15072.4 ACPB03017604 AB901066 BAQ02366.1 KQ414667 KOC64832.1 AJWK01009124 GL447257 EFN86631.1 KQ980581 KYN15288.1 ADMH02001962 ETN60224.1 KQ982373 KYQ56988.1 KQ434809 KZC06729.1 KQ976500 KYM83206.1 ADTU01011283 KQ435697 KOX80765.1 BABH01018387 APCN01004224 KQ981673 KYN38343.1 GL434492 EFN74848.1 KK854126 PTY11753.1 AXCM01000161 AXCN02000773 DS235430 EEB15716.1 AJVK01005422 LBMM01000140 KMR04783.1 QOIP01000004 RLU23501.1 KQ977059 KYN06045.1 ATLV01017584 KE525174 KFB42307.1 KZ288229 PBC31677.1 GL888932 EGI57211.1 KK107372 EZA51700.1 GECU01034016 JAS73690.1 GDIQ01239995 JAK11730.1 GDIP01237473 GDIP01204568 GDIP01121437 GDIP01093679 GDIP01083749 GDIP01079601 LRGB01000311 JAI85928.1 KZS19803.1 GDIQ01241535 JAK10190.1 GDIQ01053004 JAN41733.1 LNIX01000003 OXA58357.1 GDIQ01225036 JAK26689.1 GL732601 EFX72237.1 LJIJ01000174 ODN01053.1 HACA01023814 CDW41175.1 KQ784025 OAD51877.1 APGK01045776 KB741039 ENN74518.1 GDIQ01014409 JAN80328.1 KB632367 ERL93666.1 GDIP01222290 JAJ01112.1 GDRN01105627 JAI57703.1 KQ971354 KYB26540.1 OXA57021.1 NNAY01001300 OXU24432.1 GFDF01009470 JAV04614.1 GDIQ01202583 JAK49142.1 IACF01000422 LAB66198.1 KQ775154 OAD52283.1 GAMC01019093 GAMC01019092 JAB87463.1 KQ434977 KZC12919.1 KZ288203 PBC33461.1 JR039436 AEY58702.1 AAZX01001793 AJWK01000224 AJWK01000225 KQ977041 KYN06178.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000007151
UP000053268
UP000027135
+ More
UP000037510 UP000235965 UP000245037 UP000008820 UP000183832 UP000007062 UP000015103 UP000075881 UP000075920 UP000075900 UP000053825 UP000092461 UP000008237 UP000005203 UP000078492 UP000000673 UP000075809 UP000076502 UP000078540 UP000005205 UP000053105 UP000075884 UP000075840 UP000078541 UP000000311 UP000075903 UP000076407 UP000075902 UP000075882 UP000075883 UP000075885 UP000076408 UP000075886 UP000009046 UP000092462 UP000036403 UP000279307 UP000078542 UP000075880 UP000030765 UP000242457 UP000007755 UP000069272 UP000053097 UP000075901 UP000076858 UP000192223 UP000198287 UP000002358 UP000000305 UP000094527 UP000019118 UP000030742 UP000007266 UP000215335
UP000037510 UP000235965 UP000245037 UP000008820 UP000183832 UP000007062 UP000015103 UP000075881 UP000075920 UP000075900 UP000053825 UP000092461 UP000008237 UP000005203 UP000078492 UP000000673 UP000075809 UP000076502 UP000078540 UP000005205 UP000053105 UP000075884 UP000075840 UP000078541 UP000000311 UP000075903 UP000076407 UP000075902 UP000075882 UP000075883 UP000075885 UP000076408 UP000075886 UP000009046 UP000092462 UP000036403 UP000279307 UP000078542 UP000075880 UP000030765 UP000242457 UP000007755 UP000069272 UP000053097 UP000075901 UP000076858 UP000192223 UP000198287 UP000002358 UP000000305 UP000094527 UP000019118 UP000030742 UP000007266 UP000215335
Interpro
CDD
ProteinModelPortal
H9JWL2
A0A0N1IQH6
A0A2H1VAG8
A0A2A4JI40
A0A212F3X0
A0A194Q0T7
+ More
A0A2W1B6A7 A0A067RJL1 A0A2H1X014 A0A212EVB4 A0A2A4JIX3 A0A2W1BFS2 A0A0N1PK48 A0A194PZZ5 A0A0L7KX71 A0A1B6DDZ3 A0A2J7Q9G2 A0A2P8YW86 Q176C5 A0A1J1I0H3 A0A0P6IYT0 Q7PVH7 T1IEH0 A0A0A8JAF9 A0A182KBT0 A0A1S4H1F3 A0A182VUH5 A0A182RIN3 A0A0L7R1T9 A0A1B0CER3 E2BC95 A0A088A5V8 A0A195DQY8 W5JBS0 A0A151X9N0 A0A154P451 A0A195BF50 A0A158NBQ8 A0A0M9AAU7 H9JWL4 A0A182NU56 A0A182HGG5 A0A195FD64 E1ZVD8 A0A182VIP1 A0A182X0S9 A0A182U3K1 A0A2R7VW70 A0A182L6E7 A0A182MC20 A0A182PCF2 A0A182YDP8 A0A182QFL9 E0VQR0 A0A1B0DDL8 A0A1S4H0K6 A0A0J7LAB8 A0A3L8DSZ0 A0A151IMC3 A0A182J1P4 A0A084VWG3 A0A2A3EIX5 F4X880 A0A182F4Q8 A0A026W6L6 A0A182S8S4 A0A1B6HG68 A0A0N8AV33 A0A0N7ZK58 A0A1W4WQM3 A0A0P5GJM1 A0A0N8E7Y3 A0A226EMJ4 A0A0P5I1M3 K7JNF8 E9H7T9 A0A1D2N708 A0A0K2USY8 A0A310SBY7 N6T9V7 A0A0P6HV98 U4UTZ1 A0A0P4YV72 A0A0P4VZ78 A0A139WEZ8 A0A226EJU1 A0A232F1L7 A0A1L8DE67 A0A0P5JAK1 A0A2P2HWM8 A0A310SDC8 W8ASC7 A0A154PM32 A0A2A3EQZ2 V9IDH9 K7J7U8 A0A1B0C869 A0A195CZT9
A0A2W1B6A7 A0A067RJL1 A0A2H1X014 A0A212EVB4 A0A2A4JIX3 A0A2W1BFS2 A0A0N1PK48 A0A194PZZ5 A0A0L7KX71 A0A1B6DDZ3 A0A2J7Q9G2 A0A2P8YW86 Q176C5 A0A1J1I0H3 A0A0P6IYT0 Q7PVH7 T1IEH0 A0A0A8JAF9 A0A182KBT0 A0A1S4H1F3 A0A182VUH5 A0A182RIN3 A0A0L7R1T9 A0A1B0CER3 E2BC95 A0A088A5V8 A0A195DQY8 W5JBS0 A0A151X9N0 A0A154P451 A0A195BF50 A0A158NBQ8 A0A0M9AAU7 H9JWL4 A0A182NU56 A0A182HGG5 A0A195FD64 E1ZVD8 A0A182VIP1 A0A182X0S9 A0A182U3K1 A0A2R7VW70 A0A182L6E7 A0A182MC20 A0A182PCF2 A0A182YDP8 A0A182QFL9 E0VQR0 A0A1B0DDL8 A0A1S4H0K6 A0A0J7LAB8 A0A3L8DSZ0 A0A151IMC3 A0A182J1P4 A0A084VWG3 A0A2A3EIX5 F4X880 A0A182F4Q8 A0A026W6L6 A0A182S8S4 A0A1B6HG68 A0A0N8AV33 A0A0N7ZK58 A0A1W4WQM3 A0A0P5GJM1 A0A0N8E7Y3 A0A226EMJ4 A0A0P5I1M3 K7JNF8 E9H7T9 A0A1D2N708 A0A0K2USY8 A0A310SBY7 N6T9V7 A0A0P6HV98 U4UTZ1 A0A0P4YV72 A0A0P4VZ78 A0A139WEZ8 A0A226EJU1 A0A232F1L7 A0A1L8DE67 A0A0P5JAK1 A0A2P2HWM8 A0A310SDC8 W8ASC7 A0A154PM32 A0A2A3EQZ2 V9IDH9 K7J7U8 A0A1B0C869 A0A195CZT9
PDB
6H7D
E-value=6.96972e-23,
Score=266
Ontologies
GO
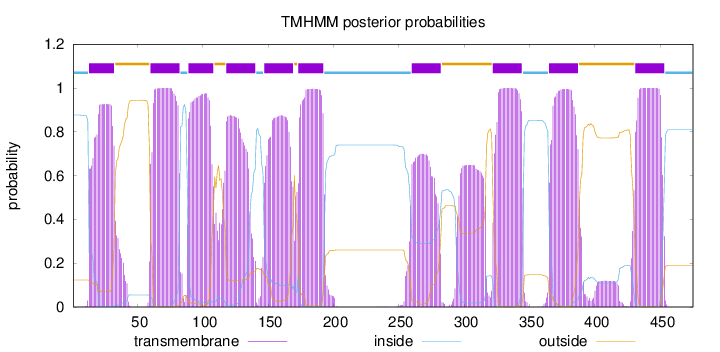
Topology
Length:
475
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
219.33534
Exp number, first 60 AAs:
19.91254
Total prob of N-in:
0.87674
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 32
outside
33 - 59
TMhelix
60 - 82
inside
83 - 88
TMhelix
89 - 108
outside
109 - 117
TMhelix
118 - 140
inside
141 - 146
TMhelix
147 - 169
outside
170 - 172
TMhelix
173 - 192
inside
193 - 259
TMhelix
260 - 282
outside
283 - 321
TMhelix
322 - 344
inside
345 - 364
TMhelix
365 - 387
outside
388 - 430
TMhelix
431 - 453
inside
454 - 475
Population Genetic Test Statistics
Pi
207.347897
Theta
149.501751
Tajima's D
1.160975
CLR
0.197465
CSRT
0.706514674266287
Interpretation
Uncertain
