Gene
KWMTBOMO16188
Pre Gene Modal
BGIBMGA013928
Annotation
PREDICTED:_facilitated_trehalose_transporter_Tret1-like_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 4.412
Sequence
CDS
ATGGCGGGCTCGAGGGAGCGTCTCCTTTACGAGAGGAAGCCTGATTACGACTCCGAATACTATCGTCAAAGATTCGAGCTACACGATCGGGAATCGAACAGGAATCGGCAGGAACAAAAGCCGCCGCGCTACTACAGACCGAAACGTACCAACGGGTACGTCTACGAAGACATATACTCGGACTTCGACGAGGCGCAGGGCCGGCTGTCCCTGTTCAGGCCAATCAGACCGGAGTACCTAAACCTGCCGACTTCGTATGACGTTAAATACAATTCCCTACCCCGAAACTACTTCGATTACGACGAAAGACGAAGAAACAGGAAAGACAATTACGATTTTCGAATCCATAACGAGAGGAGGAATGCCGAGTATTACCGGAAAAGGCCGAGAGAGGTCGTGAGCGAAATACGAAATTCAAAGCCCAAGAACGTGGCGGTCTGCAAGCCGTTACGTTTCGAAAAGAACGCTCGGTATTGGACCACGGACCCGCCCGAGCGCGGCCGAACACGCCCGAAGAAACCCGACGAACCAAAGAAGAACGTTAAGTTCTCCGAAGTTTCCGGCTGCAATCGTAAAATGATCGTAGACGACCCGATCGTAGGCCGGAAAATCGTCCCAGTGCGCGAACTCGAAGTGTCTCTAGACCAAATGATTCAGAGTGGATATTTCGAGAAGCATAACATTCCTATATCTAGGCCGTTGAAAGATGCAGAGGAAAAGATAAGCGCGTTGGAGCCGACGAGAGTGGTGCCAAACGGGAAGTGTTTATCGGACCAGCCCAGGAACGAAAATGTCGCAGTCAGGAAGACTAGGCATTTACCGCAGGTCTTGGCAGCCCTTGCTGTGTCTCTGGCGCCATTCTCAGCTGGTCTCGGTAAGGGCTACAGTTCTCCGGCCATCGCTTCTCTTCAAGGACCTGGAGCCAATGCCACCAGAAGGGATTTCCAGTTGACCGACCAGCAGGCGAGCTGGCTAGCATCACTGTCGTTTCTTGGGGCGCTCTTCGGTGGAATGGCCGGTGGTGCCGCCATGAGGCACGGGAGACGCCGCGTCCTCTCATTAGCAGCAGCGCCCTGCAGTCTATCCTGGTTGCTGACAGTACTGGCTACGTCAGTCAGGATGATGTGCATTACTGCCTTCCTGGGAGGGTTTTGCTGCTCCATACTAACCATGCTATCTCAGGTGTACATCAGTGAAATCTCAGTGCCTGACATAAGAGGATGTCTGAGCGCGGTTTTGAAGATCGTCGGCCATCTTGGAGTACTCTTTAGCTTCACTATAGGAGCGTATTTGGATTGGCAACAATTAGCTCTGTGCATTTCTGCTGCTCCCCTACTACTTTTTTGCACAGTTCTATACATCCCGGAGACCCCCAGCTACCTTGTTCTGATCGGTAAAGACGAAGAAGCTTACAAAAGTCTTCTATGGCTACGAGGTCCTGATTCAGATGTTGCACAGGAGCTAGCCACGATTCGTACTAACGTGCTGGCTAGCAAGAATTTCAGTCAACGCCACCAAATATCCAATGGTCAATTGTTTAGTTCTCTTGACGCTAGGACCATGAATCGGTTGCTTGGGCCCATCATTGTGACCTGCGGTCTGATGATGTTCCAAAGACTGTCTGGAGCCCATGCATTCTCGTTCTACGCTGTACCGATTTTCAGGAAGACATTTGGAGGTATGAACCCACACGGAGCGGCCATTGCTGTGTCCTTTGTCCAATTACTGGCCTCTTGTCTCTCTGGGCTGCTCATCGATACTGTGGGTAGGCTGCCACTGTTAATACTCAGTTCTGTGATGATGTCAATGGCATTAGCCGGCTTCGGGAGTTACGCTTACTATGAAGAGGTCCACCGTAACCAGAGAATACAAAGCGTTATGTTCCATCAGACCGATGGTCAAAACGATTGGATCCCGCTTTTATGCGTTCTGGTGTTCACAATTGCCTTCTCACTAGGAATGAGTCCTATATCATGGTTGTTAATCGGGGAATTATTTCCCCTCGAATACCGGGCATTTGGGAGCGCAATGGCAACAGCTTTTAGCTATCTGTGCGCGTTCGTAGGAGTCAAGACCTTTGTGGATTTCCAACAGGCGCTAGGACTCCATGGGGCATTCTGGTTATACGCATCAATTAGTGTGGGGGGATTGTGTTTTGTGGTCTGTTGCGTGCCGGAGACTAAAGGGAAGGATCTCGATGAAATGGACCCGAATTATGTACAAAGTTTGTCCCCTAAAAGATGTGAACTAGGTATATAA
Protein
MAGSRERLLYERKPDYDSEYYRQRFELHDRESNRNRQEQKPPRYYRPKRTNGYVYEDIYSDFDEAQGRLSLFRPIRPEYLNLPTSYDVKYNSLPRNYFDYDERRRNRKDNYDFRIHNERRNAEYYRKRPREVVSEIRNSKPKNVAVCKPLRFEKNARYWTTDPPERGRTRPKKPDEPKKNVKFSEVSGCNRKMIVDDPIVGRKIVPVRELEVSLDQMIQSGYFEKHNIPISRPLKDAEEKISALEPTRVVPNGKCLSDQPRNENVAVRKTRHLPQVLAALAVSLAPFSAGLGKGYSSPAIASLQGPGANATRRDFQLTDQQASWLASLSFLGALFGGMAGGAAMRHGRRRVLSLAAAPCSLSWLLTVLATSVRMMCITAFLGGFCCSILTMLSQVYISEISVPDIRGCLSAVLKIVGHLGVLFSFTIGAYLDWQQLALCISAAPLLLFCTVLYIPETPSYLVLIGKDEEAYKSLLWLRGPDSDVAQELATIRTNVLASKNFSQRHQISNGQLFSSLDARTMNRLLGPIIVTCGLMMFQRLSGAHAFSFYAVPIFRKTFGGMNPHGAAIAVSFVQLLASCLSGLLIDTVGRLPLLILSSVMMSMALAGFGSYAYYEEVHRNQRIQSVMFHQTDGQNDWIPLLCVLVFTIAFSLGMSPISWLLIGELFPLEYRAFGSAMATAFSYLCAFVGVKTFVDFQQALGLHGAFWLYASISVGGLCFVVCCVPETKGKDLDEMDPNYVQSLSPKRCELGI
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
EMBL
KZ150284
PZC71620.1
NWSH01001318
PCG71726.1
KQ459630
KPJ20528.1
+ More
KQ459583 KPI98608.1 AXCN02000773 ATLV01017584 KE525174 KFB42307.1 UFQS01000036 UFQT01000036 SSW97972.1 SSX18358.1 GDIP01237473 GDIP01204568 GDIP01121437 GDIP01093679 GDIP01083749 GDIP01079601 LRGB01000311 JAI85928.1 KZS19803.1 GDIQ01239995 JAK11730.1 GDIQ01241535 JAK10190.1 GDIQ01053004 JAN41733.1
KQ459583 KPI98608.1 AXCN02000773 ATLV01017584 KE525174 KFB42307.1 UFQS01000036 UFQT01000036 SSW97972.1 SSX18358.1 GDIP01237473 GDIP01204568 GDIP01121437 GDIP01093679 GDIP01083749 GDIP01079601 LRGB01000311 JAI85928.1 KZS19803.1 GDIQ01239995 JAK11730.1 GDIQ01241535 JAK10190.1 GDIQ01053004 JAN41733.1
Proteomes
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
PDB
6N3I
E-value=1.50943e-25,
Score=291
Ontologies
GO
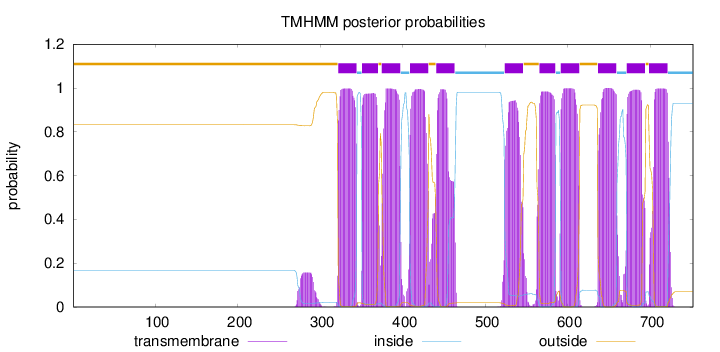
Topology
Length:
752
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
241.78698
Exp number, first 60 AAs:
0
Total prob of N-in:
0.16793
outside
1 - 321
TMhelix
322 - 344
inside
345 - 350
TMhelix
351 - 370
outside
371 - 374
TMhelix
375 - 397
inside
398 - 408
TMhelix
409 - 431
outside
432 - 440
TMhelix
441 - 463
inside
464 - 523
TMhelix
524 - 546
outside
547 - 565
TMhelix
566 - 585
inside
586 - 591
TMhelix
592 - 614
outside
615 - 636
TMhelix
637 - 659
inside
660 - 671
TMhelix
672 - 694
outside
695 - 698
TMhelix
699 - 721
inside
722 - 752
Population Genetic Test Statistics
Pi
204.523933
Theta
153.982713
Tajima's D
0.982291
CLR
0.522704
CSRT
0.664966751662417
Interpretation
Uncertain
