Gene
KWMTBOMO16185
Annotation
PREDICTED:_uncharacterized_protein_LOC106126082_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 3.835
Sequence
CDS
ATGGTTCACTGCTCGGAGTGTAAACTGAACATTTTAGTCACCCAGAGGCGAATCAAATGCACTAATCCGGACTGCCAACACAATTACCATCAGGACTGTGTCAAGTACAATGATACTACCACCGCTCGCTCTAAATGGATTTGCCCCGATTGTACAATACCGCGCCAAAAAAGTGGAAATACACCTCTTTCCATGTCGAAGAATAAAGACATTTCGGATGCTTCTGCACCCTTATTGGGAAGTCAAGAATTAATTCTATTAGAAATACGTAATCTACGCCAAGAAATGTTGGACAAATTTGAGTGCCAGAAATCGGCGTTCGAGAGATTTAATGCAATTTTGTGCAAGGTTCAAATGGATATGAGTGAGTTGGACTCCAAATGCTCCGGTCTGCGAGAGGATCTCAATGGTGCTATAAATACATTGCAATTTCTATCTGCATGTCACGACGATCAGCAAAAATTGAATGAAAAAACAAAAAGTGACTTTTTGGAATTCACTAAGTGCAGCAACAGCCTCCAACCAAAACTATCAGAATTGAACCATAAAATCGCGTATATGGAGCAGTTCGCAAGACAGTCAAACGTTGAAATTCAGGGGATTCCAGAAAGACGCAATGAAAACTTAATGTCTATAGTTAAAAAAATTGGAAATACGGTTTCCTGTAATTTACAAGAAACTGACATTCTGGAATTCCACAGAGTTGCTAAGCAAAATACCAATTCGAATCGTCCGAAAAATATTGTAGTGAAGTTAAACTGCCCACGTACTCGAGACAATCTCCTTGCTTCAGTTAAAATCTTTAATAAAAACAAGCACAGTGATAACAAACTTAACACTTCCCATATTGAACTTCCTGGTGAACAGCGGCCTGTGTATGTTACTGAACACTTGTCACCTGAAAATAAAAAACTTCATGCCGCTACTAGAATAGCTGCCAAGGAGAAGAATTACAACTTTGTTTGGATACGAAATGGTCGCATCTTCGTACGTAAGGATATTTCTGCCGGCCACATTAATATTAAGACTGTAGAATGCTTGCATAAGCTGTGA
Protein
MVHCSECKLNILVTQRRIKCTNPDCQHNYHQDCVKYNDTTTARSKWICPDCTIPRQKSGNTPLSMSKNKDISDASAPLLGSQELILLEIRNLRQEMLDKFECQKSAFERFNAILCKVQMDMSELDSKCSGLREDLNGAINTLQFLSACHDDQQKLNEKTKSDFLEFTKCSNSLQPKLSELNHKIAYMEQFARQSNVEIQGIPERRNENLMSIVKKIGNTVSCNLQETDILEFHRVAKQNTNSNRPKNIVVKLNCPRTRDNLLASVKIFNKNKHSDNKLNTSHIELPGEQRPVYVTEHLSPENKKLHAATRIAAKEKNYNFVWIRNGRIFVRKDISAGHINIKTVECLHKL
Summary
Similarity
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Uniprot
A0A2A4IU35
A0A2H1W5G4
A0A2A4IXU4
A0A2W1BYU6
A0A2H1VD37
A0A2H1W117
+ More
A0A2A4JBL7 A0A2H1WBH0 A0A0L7KQ92 A0A0L7K4K4 A0A0L7L3E9 A0A2A4IW47 A0A2H1W956 A0A0L7LEU2 A0A0L7L1J8 A0A0L7LEV4 A0A0L7K3N5 A0A2A4IZ89 A0A0L7L0X6 A0A2H1VJZ0 A0A212EWC1 A0A0L7K3E6 A0A2A4IYB9 A0A2A4JC78 A0A2H1V294 A0A2A4JD31 A0A2H1X1K9 G3E4N3 A0A2A4IYM6 A0A1Y1JXQ1 A0A1S3DD01 A0A0L7K2E3 A0A0L7LF48 A0A2H1X1G7 A0A2H1WRH7 A0A0L7LRU8 A0A1E1WIH3 A0A2A4JL66 A0A2H1WVR8 A0A2A4JWA7 A0A0L7LCM2 A0A2A4JK29 A0A2H1WIH0 A0A1B6L4T1 A0A0L7L7A2 A0A2H1WQP2 A0A212F6C5 H9JHN2 A0A0L7LU57 A0A0L7L026 A0A0L7LAT0 A0A2H1V8K6 A0A0L7KJ36
A0A2A4JBL7 A0A2H1WBH0 A0A0L7KQ92 A0A0L7K4K4 A0A0L7L3E9 A0A2A4IW47 A0A2H1W956 A0A0L7LEU2 A0A0L7L1J8 A0A0L7LEV4 A0A0L7K3N5 A0A2A4IZ89 A0A0L7L0X6 A0A2H1VJZ0 A0A212EWC1 A0A0L7K3E6 A0A2A4IYB9 A0A2A4JC78 A0A2H1V294 A0A2A4JD31 A0A2H1X1K9 G3E4N3 A0A2A4IYM6 A0A1Y1JXQ1 A0A1S3DD01 A0A0L7K2E3 A0A0L7LF48 A0A2H1X1G7 A0A2H1WRH7 A0A0L7LRU8 A0A1E1WIH3 A0A2A4JL66 A0A2H1WVR8 A0A2A4JWA7 A0A0L7LCM2 A0A2A4JK29 A0A2H1WIH0 A0A1B6L4T1 A0A0L7L7A2 A0A2H1WQP2 A0A212F6C5 H9JHN2 A0A0L7LU57 A0A0L7L026 A0A0L7LAT0 A0A2H1V8K6 A0A0L7KJ36
EMBL
NWSH01007563
PCG62908.1
ODYU01006462
SOQ48321.1
NWSH01005418
PCG64228.1
+ More
KZ149914 PZC78000.1 ODYU01001881 SOQ38706.1 ODYU01005672 SOQ46768.1 NWSH01002191 PCG68944.1 ODYU01007238 SOQ49844.1 JTDY01007401 KOB65231.1 JTDY01010989 KOB57787.1 JTDY01003264 KOB69846.1 NWSH01005702 PCG64035.1 ODYU01007128 SOQ49629.1 JTDY01001361 KOB74078.1 JTDY01003535 KOB69383.1 JTDY01001411 KOB73935.1 JTDY01012405 KOB52279.1 NWSH01004298 PCG65247.1 JTDY01003733 KOB69075.1 ODYU01002971 SOQ41143.1 AGBW02012032 OWR45776.1 JTDY01014577 KOB52017.1 NWSH01005479 PCG64170.1 NWSH01002032 PCG69366.1 ODYU01000349 SOQ34960.1 NWSH01002051 PCG69303.1 ODYU01012701 SOQ59142.1 HQ585015 AEM44816.1 NWSH01004492 PCG65047.1 GEZM01097808 JAV54094.1 JTDY01015045 KOB51976.1 JTDY01001455 KOB73826.1 ODYU01012716 SOQ59161.1 ODYU01010507 SOQ55661.1 JTDY01000280 KOB77936.1 GDQN01004260 JAT86794.1 NWSH01001083 PCG72711.1 ODYU01011375 SOQ57082.1 NWSH01000452 NWSH01000043 PCG76317.1 PCG80309.1 JTDY01001653 KOB73242.1 NWSH01001156 PCG72321.1 ODYU01008890 SOQ52873.1 GEBQ01021483 JAT18494.1 JTDY01006475 JTDY01002579 KOB65982.1 KOB71161.1 ODYU01010286 SOQ55282.1 AGBW02010051 OWR49283.1 BABH01012931 BABH01012932 JTDY01000093 KOB78914.1 JTDY01003969 KOB68765.1 JTDY01001900 KOB72582.1 ODYU01001225 SOQ37147.1 JTDY01009562 KOB63342.1
KZ149914 PZC78000.1 ODYU01001881 SOQ38706.1 ODYU01005672 SOQ46768.1 NWSH01002191 PCG68944.1 ODYU01007238 SOQ49844.1 JTDY01007401 KOB65231.1 JTDY01010989 KOB57787.1 JTDY01003264 KOB69846.1 NWSH01005702 PCG64035.1 ODYU01007128 SOQ49629.1 JTDY01001361 KOB74078.1 JTDY01003535 KOB69383.1 JTDY01001411 KOB73935.1 JTDY01012405 KOB52279.1 NWSH01004298 PCG65247.1 JTDY01003733 KOB69075.1 ODYU01002971 SOQ41143.1 AGBW02012032 OWR45776.1 JTDY01014577 KOB52017.1 NWSH01005479 PCG64170.1 NWSH01002032 PCG69366.1 ODYU01000349 SOQ34960.1 NWSH01002051 PCG69303.1 ODYU01012701 SOQ59142.1 HQ585015 AEM44816.1 NWSH01004492 PCG65047.1 GEZM01097808 JAV54094.1 JTDY01015045 KOB51976.1 JTDY01001455 KOB73826.1 ODYU01012716 SOQ59161.1 ODYU01010507 SOQ55661.1 JTDY01000280 KOB77936.1 GDQN01004260 JAT86794.1 NWSH01001083 PCG72711.1 ODYU01011375 SOQ57082.1 NWSH01000452 NWSH01000043 PCG76317.1 PCG80309.1 JTDY01001653 KOB73242.1 NWSH01001156 PCG72321.1 ODYU01008890 SOQ52873.1 GEBQ01021483 JAT18494.1 JTDY01006475 JTDY01002579 KOB65982.1 KOB71161.1 ODYU01010286 SOQ55282.1 AGBW02010051 OWR49283.1 BABH01012931 BABH01012932 JTDY01000093 KOB78914.1 JTDY01003969 KOB68765.1 JTDY01001900 KOB72582.1 ODYU01001225 SOQ37147.1 JTDY01009562 KOB63342.1
Proteomes
PRIDE
Interpro
IPR004244
Transposase_22
+ More
IPR004941 Baculo_FP
IPR011011 Znf_FYVE_PHD
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR036691 Endo/exonu/phosph_ase_sf
IPR019786 Zinc_finger_PHD-type_CS
IPR019787 Znf_PHD-finger
IPR002067 Mit_carrier
IPR018108 Mitochondrial_sb/sol_carrier
IPR023395 Mt_carrier_dom_sf
IPR001841 Znf_RING
IPR005135 Endo/exonuclease/phosphatase
IPR004941 Baculo_FP
IPR011011 Znf_FYVE_PHD
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR036691 Endo/exonu/phosph_ase_sf
IPR019786 Zinc_finger_PHD-type_CS
IPR019787 Znf_PHD-finger
IPR002067 Mit_carrier
IPR018108 Mitochondrial_sb/sol_carrier
IPR023395 Mt_carrier_dom_sf
IPR001841 Znf_RING
IPR005135 Endo/exonuclease/phosphatase
Gene 3D
ProteinModelPortal
A0A2A4IU35
A0A2H1W5G4
A0A2A4IXU4
A0A2W1BYU6
A0A2H1VD37
A0A2H1W117
+ More
A0A2A4JBL7 A0A2H1WBH0 A0A0L7KQ92 A0A0L7K4K4 A0A0L7L3E9 A0A2A4IW47 A0A2H1W956 A0A0L7LEU2 A0A0L7L1J8 A0A0L7LEV4 A0A0L7K3N5 A0A2A4IZ89 A0A0L7L0X6 A0A2H1VJZ0 A0A212EWC1 A0A0L7K3E6 A0A2A4IYB9 A0A2A4JC78 A0A2H1V294 A0A2A4JD31 A0A2H1X1K9 G3E4N3 A0A2A4IYM6 A0A1Y1JXQ1 A0A1S3DD01 A0A0L7K2E3 A0A0L7LF48 A0A2H1X1G7 A0A2H1WRH7 A0A0L7LRU8 A0A1E1WIH3 A0A2A4JL66 A0A2H1WVR8 A0A2A4JWA7 A0A0L7LCM2 A0A2A4JK29 A0A2H1WIH0 A0A1B6L4T1 A0A0L7L7A2 A0A2H1WQP2 A0A212F6C5 H9JHN2 A0A0L7LU57 A0A0L7L026 A0A0L7LAT0 A0A2H1V8K6 A0A0L7KJ36
A0A2A4JBL7 A0A2H1WBH0 A0A0L7KQ92 A0A0L7K4K4 A0A0L7L3E9 A0A2A4IW47 A0A2H1W956 A0A0L7LEU2 A0A0L7L1J8 A0A0L7LEV4 A0A0L7K3N5 A0A2A4IZ89 A0A0L7L0X6 A0A2H1VJZ0 A0A212EWC1 A0A0L7K3E6 A0A2A4IYB9 A0A2A4JC78 A0A2H1V294 A0A2A4JD31 A0A2H1X1K9 G3E4N3 A0A2A4IYM6 A0A1Y1JXQ1 A0A1S3DD01 A0A0L7K2E3 A0A0L7LF48 A0A2H1X1G7 A0A2H1WRH7 A0A0L7LRU8 A0A1E1WIH3 A0A2A4JL66 A0A2H1WVR8 A0A2A4JWA7 A0A0L7LCM2 A0A2A4JK29 A0A2H1WIH0 A0A1B6L4T1 A0A0L7L7A2 A0A2H1WQP2 A0A212F6C5 H9JHN2 A0A0L7LU57 A0A0L7L026 A0A0L7LAT0 A0A2H1V8K6 A0A0L7KJ36
PDB
2K1J
E-value=0.0343485,
Score=86
Ontologies
KEGG
GO
PANTHER
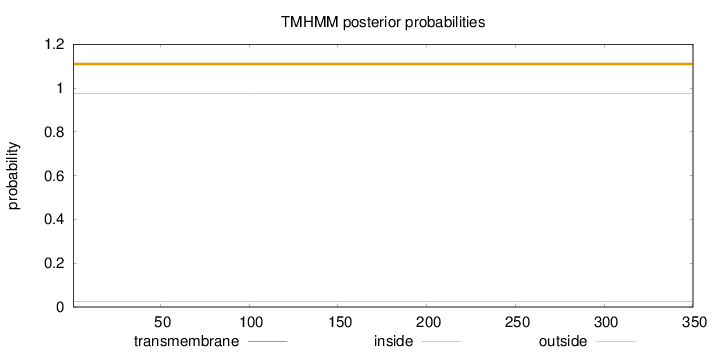
Topology
Length:
350
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00013
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02608
outside
1 - 350
Population Genetic Test Statistics
Pi
266.361161
Theta
163.887297
Tajima's D
1.966436
CLR
0
CSRT
0.874906254687266
Interpretation
Uncertain
