Gene
KWMTBOMO16184
Pre Gene Modal
BGIBMGA013929
Annotation
juvenile_hormone_epoxide_hydrolase-like_protein_5_[Bombyx_mori]
Full name
Epoxide hydrolase
+ More
Juvenile hormone epoxide hydrolase
Glutathione peroxidase
Juvenile hormone epoxide hydrolase 1
Juvenile hormone epoxide hydrolase 2
Juvenile hormone epoxide hydrolase
Glutathione peroxidase
Juvenile hormone epoxide hydrolase 1
Juvenile hormone epoxide hydrolase 2
Alternative Name
CfEH1
Juvenile hormone epoxide hydrolase I
Juvenile hormone-specific epoxide hydrolase I
CfEH2
Juvenile hormone epoxide hydrolase II
Juvenile hormone-specific epoxide hydrolase II
Juvenile hormone epoxide hydrolase I
Juvenile hormone-specific epoxide hydrolase I
CfEH2
Juvenile hormone epoxide hydrolase II
Juvenile hormone-specific epoxide hydrolase II
Location in the cell
Mitochondrial Reliability : 0.979
Sequence
CDS
ATGCGGTACGGTGTAATACAAAAGTTGAAACCGAAAGAATTCTATCCGATCCGAAATGACGTAATCGACGATTTGAAATATCGTCTCAAGAATCATCGACCCTTCGCCCCGCCACTAGAGGGTGTGACCTTCGAATACGGCTTCAACACGGCTGCCCTAGAACCCTGGCTGAAATACTGGGCCGAGGAGTATAAATTCGCTGACCGAGAAAAGTTCTTTAATCAGTTTCCACATTTCAAGACCAGCATCAAGGGCCTGGATATTCATTTCATCAGAGTTAAGCCGCAGGTATCTGCAGGAGTCCAGACTCTGCCCCTTCTTCTGTTACATGGCTGGCCAGGTTCCGTCAGAGAGTTCTATGAAGCCATCCCCCTGCTGACCAGTCAAAATTCTGGGTATGATTTTGCCTTTGAAGTCATAGTCCCGAGTCTACCCGGATATGGTTTTTCTGATGCTGCAGTGAGACCAGGCTTATCTTTGCCGTACGTAGCAGATATCTTCAGAATATTAATGAAAAGATTGGGTCATGATAAATTTTACATACAAGGGGGAGATTGGGGCGCAGCCATTGCCTCGGCCATGGTTACCCTATTCCCTGAAGATGTTCTTGGACATCACAGTAATTCAGCTGTTACACAGCATCCTCACGCCCTGCTCAGAACTCTGCTTGGTGCTTTAATACCGTCACTAATAGTGGAAGATCATCTAGCCGAAAGAATGTATCCACTTTCAAAACATCTGGCGTACTTGTTGGAAGAATTCGGATACTTTCACTTACAAGCCACAAAGCCTGATACAGTAGGTGTTTCCCTGACGGATTCTCCATCAGGTCTTCTAGCGTATATTTTAGAGAAATTCGCGGTTTGGACGCGCAAGGAGCACAAGTTTAAGAGTGACGGCGGTCTCGGCTTCAGGTTTAGCAAGGAAAAGCTGATAGATAATCTAATGGTCTACTGGATTACCAACTCTATTACAAGCTCAATGAGATTCTATTCCGAAAACATGAGTAACAAATTCAGGGAAATGAATTTGGAAGCTTTTACATCACCCGTTCCCACGTGGGCTCTCCAGGCCAGAGACGAGCTGGTCTATCAGCCGCCTGCAGTTTTACGTGCGAAGTACCTGAATCTCCTCAACGTGACGGTTTTAGACGACGGCGGACATTTCTTGGCTTTGGAACTTCCTGAGGTGTTCGCGGAAGACGTTTTCAAGGCGGTTCACGCGTTCAAGCAGTGGCGTTCTACTCATTCCAAGACTGATCATTAG
Protein
MRYGVIQKLKPKEFYPIRNDVIDDLKYRLKNHRPFAPPLEGVTFEYGFNTAALEPWLKYWAEEYKFADREKFFNQFPHFKTSIKGLDIHFIRVKPQVSAGVQTLPLLLLHGWPGSVREFYEAIPLLTSQNSGYDFAFEVIVPSLPGYGFSDAAVRPGLSLPYVADIFRILMKRLGHDKFYIQGGDWGAAIASAMVTLFPEDVLGHHSNSAVTQHPHALLRTLLGALIPSLIVEDHLAERMYPLSKHLAYLLEEFGYFHLQATKPDTVGVSLTDSPSGLLAYILEKFAVWTRKEHKFKSDGGLGFRFSKEKLIDNLMVYWITNSITSSMRFYSENMSNKFREMNLEAFTSPVPTWALQARDELVYQPPAVLRAKYLNLLNVTVLDDGGHFLALELPEVFAEDVFKAVHAFKQWRSTHSKTDH
Summary
Description
Catalyzes juvenile hormone hydrolysis.
Catalyzes juvenile hormone hydrolysis. Degrades juvenile hormone III (JH III) about 3 times and 5 times slower than juvenile hormone I (JH I) and II (JH II), respectively. Degrades cis-stilbene oxide and trans-stilbene oxide about 18 and 43 times slower than JH III, respectively.
Catalyzes juvenile hormone hydrolysis. Degrades juvenile hormone III (JH III) about 3 times and 5 times slower than juvenile hormone I (JH I) and II (JH II), respectively. Degrades cis-stilbene oxide and trans-stilbene oxide about 18 and 43 times slower than JH III, respectively.
Catalytic Activity
1-(4-methoxyphenyl)-N-methyl-N-[(3-methyloxetan-3-yl)methyl]methanamine + H2O = 2-{[(4-methoxybenzyl)(methyl)amino]methyl}-2-methylpropane-1,3-diol
cis-stilbene oxide + H2O = (1R,2R)-hydrobenzoin
cis-stilbene oxide + H2O = (1R,2R)-hydrobenzoin
Biophysicochemical Properties
0.52 uM for juvenile hormone III
Subunit
Homodimer.
Similarity
Belongs to the peptidase S33 family.
Belongs to the glutathione peroxidase family.
Belongs to the glutathione peroxidase family.
Keywords
3D-structure
Aromatic hydrocarbons catabolism
Complete proteome
Endoplasmic reticulum
Hydrolase
Membrane
Microsome
Reference proteome
Transmembrane
Transmembrane helix
Direct protein sequencing
Feature
chain Epoxide hydrolase
Uniprot
D4Q9H4
H9JWL5
Q1W696
A0A2W1B2C5
A0A194Q5D4
A0A2A4K9D8
+ More
L7R9X8 A0A194PZ48 A0A0N1IBG8 A0A3S2P9A9 A0A0R6CEE9 A0A2A4K8F5 A0A0N1IQA6 A0A2W1B6Y8 C0KH33 I4DKH4 A0A194PZ44 A0A212F9X4 A0A3S2LEJ8 A0A0N1ICQ7 H9JWL6 Q6U6J0 Q94806 O44124 A0A0N1IIM0 A0A194PZ55 A0A212F9Y7 A0A212F9W9 A0A194Q0T4 A0A0N0PEI4 A0A2A4JI09 A0A2W1B693 A0A0N0PCA1 C6KWM6 A0A1Y1K079 A0A1Y1K345 A0A2A4J6Z6 A0A0T6B3Y6 A0A088M9L0 A0A0T6BH94 D0W014 A0A0N0PD06 A0A2W1BCV7 A0A2A4JKY8 A0A194PZZ0 D6WGJ4 A0A2C9DK11 A0A2J7PHG7 H9JWT0 H9JI63 Q8MZR6 C6KWM7 A0A2H1VS44 V5G055 A0A067QLM6 D0W015 D6WGJ2 F4WV40 R4G4K1 A0A0P0ELB6 A0A154PRH0 A0A3S2NYE4 A0A226E0H1 D0W012 A0A1W4XE43 A0A0F6SIA8 J3JY41 A0A195EXA2 A0A1V0M8A5 A0A195DS73 Q8MZR5 D0W011 A0A0T6BDK1 B3U0E6 D6WGJ1 A0A0A9WSP4 V5G602 A0A3G6VF49 A0A0K8SHU3 A0A1S4EFE4 A0A151WUE8 A0A0N0PAA2 A0A3S2NQU9 N6UPP4 A0A146KNU3 A0A087ZVE8 A0A151I9X6 A0A195CR36 A0A1W6BRF0 A0A224XNL2 A0A2D3FB68 A0A0V0G516 A0A1W5KJY8 A0A0T6BDP9 A0A0L7QRY7 A0A195DZH4 A0A2A3E8M7 A0A069DUW8 A0A2P8YCD2 A0A158NNQ4
L7R9X8 A0A194PZ48 A0A0N1IBG8 A0A3S2P9A9 A0A0R6CEE9 A0A2A4K8F5 A0A0N1IQA6 A0A2W1B6Y8 C0KH33 I4DKH4 A0A194PZ44 A0A212F9X4 A0A3S2LEJ8 A0A0N1ICQ7 H9JWL6 Q6U6J0 Q94806 O44124 A0A0N1IIM0 A0A194PZ55 A0A212F9Y7 A0A212F9W9 A0A194Q0T4 A0A0N0PEI4 A0A2A4JI09 A0A2W1B693 A0A0N0PCA1 C6KWM6 A0A1Y1K079 A0A1Y1K345 A0A2A4J6Z6 A0A0T6B3Y6 A0A088M9L0 A0A0T6BH94 D0W014 A0A0N0PD06 A0A2W1BCV7 A0A2A4JKY8 A0A194PZZ0 D6WGJ4 A0A2C9DK11 A0A2J7PHG7 H9JWT0 H9JI63 Q8MZR6 C6KWM7 A0A2H1VS44 V5G055 A0A067QLM6 D0W015 D6WGJ2 F4WV40 R4G4K1 A0A0P0ELB6 A0A154PRH0 A0A3S2NYE4 A0A226E0H1 D0W012 A0A1W4XE43 A0A0F6SIA8 J3JY41 A0A195EXA2 A0A1V0M8A5 A0A195DS73 Q8MZR5 D0W011 A0A0T6BDK1 B3U0E6 D6WGJ1 A0A0A9WSP4 V5G602 A0A3G6VF49 A0A0K8SHU3 A0A1S4EFE4 A0A151WUE8 A0A0N0PAA2 A0A3S2NQU9 N6UPP4 A0A146KNU3 A0A087ZVE8 A0A151I9X6 A0A195CR36 A0A1W6BRF0 A0A224XNL2 A0A2D3FB68 A0A0V0G516 A0A1W5KJY8 A0A0T6BDP9 A0A0L7QRY7 A0A195DZH4 A0A2A3E8M7 A0A069DUW8 A0A2P8YCD2 A0A158NNQ4
EC Number
3.3.2.9
Pubmed
EMBL
AB299279
BAI94514.1
BABH01018381
BABH01018382
DQ431847
ABD85119.1
+ More
KZ150418 PZC70942.1 KQ459583 KPI98605.1 NWSH01000031 PCG80508.1 JX681816 AGC00788.1 KPI98607.1 KQ459630 KPJ20526.1 RSAL01000177 RVE45007.1 KM926340 AJR27470.1 PCG80507.1 KPJ20527.1 PZC70941.1 FJ602793 ACM78602.2 AK401792 BAM18414.1 KPI98602.1 AGBW02009550 OWR50537.1 RVE45005.1 KQ459764 KPJ20092.1 BABH01018379 AY377854 AB362775 AAQ87024.1 BAF81491.1 U73680 AAB18243.1 AF035482 AAB88192.1 KPJ20091.1 KPI98601.1 OWR50538.1 OWR50536.1 KPI98604.1 KPJ20089.1 NWSH01001318 PCG71725.1 KZ150284 PZC70943.1 PZC71622.1 KQ460733 KPJ12641.1 AB293555 BAH97089.1 GEZM01096421 JAV54922.1 GEZM01096422 JAV54921.1 NWSH01002656 PCG67731.1 LJIG01009934 KRT82088.1 KJ579233 AIN34709.1 LJIG01000290 KRT86650.1 AB534188 BAI49689.1 KQ460417 KPJ14867.1 KZ150476 PZC70740.1 NWSH01001212 PCG72103.1 KPI98603.1 KQ971321 EFA00569.1 KX855995 ARE68677.1 NEVH01025136 PNF15772.1 BABH01018608 BABH01022402 AF503908 AAM22694.1 AB293556 BAH97090.1 ODYU01004100 SOQ43618.1 GALX01005126 JAB63340.1 KK853182 KDR10172.1 AB534189 BAI49690.1 EFA00568.1 GL888384 EGI61928.1 ACPB03008739 GAHY01001290 JAA76220.1 KT261710 ALJ30250.1 KQ435007 KZC13710.1 RSAL01000025 RVE52162.1 LNIX01000009 OXA50467.1 AB534186 BAI49687.1 KP271046 AKF11871.1 BT128165 AEE63126.1 KQ981953 KYN32509.1 KU755500 ARD71210.1 KQ980530 KYN15652.1 AF503909 AAM22695.1 AB534185 BAI49686.1 LJIG01001532 KRT85408.1 EU441215 ACC55233.1 EFA00567.1 GBHO01035734 JAG07870.1 GALX01002976 JAB65490.1 MH932586 AZB52850.1 GBRD01012965 JAG52861.1 KQ982745 KYQ51387.1 KQ459037 KPJ04294.1 RSAL01000008 RVE53920.1 APGK01022356 APGK01022357 KB740419 ENN80707.1 GDHC01020455 JAP98173.1 KQ978260 KYM95872.1 KQ977381 KYN03166.1 KY810195 ARJ54611.1 GFTR01006406 JAW10020.1 KY407548 ATU47254.1 GECL01002991 JAP03133.1 KU941586 AMR44689.1 LJIG01001533 KRT85407.1 KQ414766 KOC61382.1 KQ979999 KYN18300.1 KZ288345 PBC27632.1 GBGD01001402 JAC87487.1 PYGN01000706 PSN41919.1 ADTU01021716
KZ150418 PZC70942.1 KQ459583 KPI98605.1 NWSH01000031 PCG80508.1 JX681816 AGC00788.1 KPI98607.1 KQ459630 KPJ20526.1 RSAL01000177 RVE45007.1 KM926340 AJR27470.1 PCG80507.1 KPJ20527.1 PZC70941.1 FJ602793 ACM78602.2 AK401792 BAM18414.1 KPI98602.1 AGBW02009550 OWR50537.1 RVE45005.1 KQ459764 KPJ20092.1 BABH01018379 AY377854 AB362775 AAQ87024.1 BAF81491.1 U73680 AAB18243.1 AF035482 AAB88192.1 KPJ20091.1 KPI98601.1 OWR50538.1 OWR50536.1 KPI98604.1 KPJ20089.1 NWSH01001318 PCG71725.1 KZ150284 PZC70943.1 PZC71622.1 KQ460733 KPJ12641.1 AB293555 BAH97089.1 GEZM01096421 JAV54922.1 GEZM01096422 JAV54921.1 NWSH01002656 PCG67731.1 LJIG01009934 KRT82088.1 KJ579233 AIN34709.1 LJIG01000290 KRT86650.1 AB534188 BAI49689.1 KQ460417 KPJ14867.1 KZ150476 PZC70740.1 NWSH01001212 PCG72103.1 KPI98603.1 KQ971321 EFA00569.1 KX855995 ARE68677.1 NEVH01025136 PNF15772.1 BABH01018608 BABH01022402 AF503908 AAM22694.1 AB293556 BAH97090.1 ODYU01004100 SOQ43618.1 GALX01005126 JAB63340.1 KK853182 KDR10172.1 AB534189 BAI49690.1 EFA00568.1 GL888384 EGI61928.1 ACPB03008739 GAHY01001290 JAA76220.1 KT261710 ALJ30250.1 KQ435007 KZC13710.1 RSAL01000025 RVE52162.1 LNIX01000009 OXA50467.1 AB534186 BAI49687.1 KP271046 AKF11871.1 BT128165 AEE63126.1 KQ981953 KYN32509.1 KU755500 ARD71210.1 KQ980530 KYN15652.1 AF503909 AAM22695.1 AB534185 BAI49686.1 LJIG01001532 KRT85408.1 EU441215 ACC55233.1 EFA00567.1 GBHO01035734 JAG07870.1 GALX01002976 JAB65490.1 MH932586 AZB52850.1 GBRD01012965 JAG52861.1 KQ982745 KYQ51387.1 KQ459037 KPJ04294.1 RSAL01000008 RVE53920.1 APGK01022356 APGK01022357 KB740419 ENN80707.1 GDHC01020455 JAP98173.1 KQ978260 KYM95872.1 KQ977381 KYN03166.1 KY810195 ARJ54611.1 GFTR01006406 JAW10020.1 KY407548 ATU47254.1 GECL01002991 JAP03133.1 KU941586 AMR44689.1 LJIG01001533 KRT85407.1 KQ414766 KOC61382.1 KQ979999 KYN18300.1 KZ288345 PBC27632.1 GBGD01001402 JAC87487.1 PYGN01000706 PSN41919.1 ADTU01021716
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000283053
UP000007151
+ More
UP000007266 UP000235965 UP000027135 UP000007755 UP000015103 UP000076502 UP000198287 UP000192223 UP000078541 UP000078492 UP000002358 UP000079169 UP000075809 UP000019118 UP000005203 UP000078542 UP000053825 UP000242457 UP000245037 UP000005205
UP000007266 UP000235965 UP000027135 UP000007755 UP000015103 UP000076502 UP000198287 UP000192223 UP000078541 UP000078492 UP000002358 UP000079169 UP000075809 UP000019118 UP000005203 UP000078542 UP000053825 UP000242457 UP000245037 UP000005205
Interpro
IPR029058
AB_hydrolase
+ More
IPR016292 Epoxide_hydrolase
IPR010497 Epoxide_hydro_N
IPR000639 Epox_hydrolase-like
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR029759 GPX_AS
IPR036249 Thioredoxin-like_sf
IPR029760 GPX_CS
IPR000889 Glutathione_peroxidase
IPR016292 Epoxide_hydrolase
IPR010497 Epoxide_hydro_N
IPR000639 Epox_hydrolase-like
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR029759 GPX_AS
IPR036249 Thioredoxin-like_sf
IPR029760 GPX_CS
IPR000889 Glutathione_peroxidase
Gene 3D
ProteinModelPortal
D4Q9H4
H9JWL5
Q1W696
A0A2W1B2C5
A0A194Q5D4
A0A2A4K9D8
+ More
L7R9X8 A0A194PZ48 A0A0N1IBG8 A0A3S2P9A9 A0A0R6CEE9 A0A2A4K8F5 A0A0N1IQA6 A0A2W1B6Y8 C0KH33 I4DKH4 A0A194PZ44 A0A212F9X4 A0A3S2LEJ8 A0A0N1ICQ7 H9JWL6 Q6U6J0 Q94806 O44124 A0A0N1IIM0 A0A194PZ55 A0A212F9Y7 A0A212F9W9 A0A194Q0T4 A0A0N0PEI4 A0A2A4JI09 A0A2W1B693 A0A0N0PCA1 C6KWM6 A0A1Y1K079 A0A1Y1K345 A0A2A4J6Z6 A0A0T6B3Y6 A0A088M9L0 A0A0T6BH94 D0W014 A0A0N0PD06 A0A2W1BCV7 A0A2A4JKY8 A0A194PZZ0 D6WGJ4 A0A2C9DK11 A0A2J7PHG7 H9JWT0 H9JI63 Q8MZR6 C6KWM7 A0A2H1VS44 V5G055 A0A067QLM6 D0W015 D6WGJ2 F4WV40 R4G4K1 A0A0P0ELB6 A0A154PRH0 A0A3S2NYE4 A0A226E0H1 D0W012 A0A1W4XE43 A0A0F6SIA8 J3JY41 A0A195EXA2 A0A1V0M8A5 A0A195DS73 Q8MZR5 D0W011 A0A0T6BDK1 B3U0E6 D6WGJ1 A0A0A9WSP4 V5G602 A0A3G6VF49 A0A0K8SHU3 A0A1S4EFE4 A0A151WUE8 A0A0N0PAA2 A0A3S2NQU9 N6UPP4 A0A146KNU3 A0A087ZVE8 A0A151I9X6 A0A195CR36 A0A1W6BRF0 A0A224XNL2 A0A2D3FB68 A0A0V0G516 A0A1W5KJY8 A0A0T6BDP9 A0A0L7QRY7 A0A195DZH4 A0A2A3E8M7 A0A069DUW8 A0A2P8YCD2 A0A158NNQ4
L7R9X8 A0A194PZ48 A0A0N1IBG8 A0A3S2P9A9 A0A0R6CEE9 A0A2A4K8F5 A0A0N1IQA6 A0A2W1B6Y8 C0KH33 I4DKH4 A0A194PZ44 A0A212F9X4 A0A3S2LEJ8 A0A0N1ICQ7 H9JWL6 Q6U6J0 Q94806 O44124 A0A0N1IIM0 A0A194PZ55 A0A212F9Y7 A0A212F9W9 A0A194Q0T4 A0A0N0PEI4 A0A2A4JI09 A0A2W1B693 A0A0N0PCA1 C6KWM6 A0A1Y1K079 A0A1Y1K345 A0A2A4J6Z6 A0A0T6B3Y6 A0A088M9L0 A0A0T6BH94 D0W014 A0A0N0PD06 A0A2W1BCV7 A0A2A4JKY8 A0A194PZZ0 D6WGJ4 A0A2C9DK11 A0A2J7PHG7 H9JWT0 H9JI63 Q8MZR6 C6KWM7 A0A2H1VS44 V5G055 A0A067QLM6 D0W015 D6WGJ2 F4WV40 R4G4K1 A0A0P0ELB6 A0A154PRH0 A0A3S2NYE4 A0A226E0H1 D0W012 A0A1W4XE43 A0A0F6SIA8 J3JY41 A0A195EXA2 A0A1V0M8A5 A0A195DS73 Q8MZR5 D0W011 A0A0T6BDK1 B3U0E6 D6WGJ1 A0A0A9WSP4 V5G602 A0A3G6VF49 A0A0K8SHU3 A0A1S4EFE4 A0A151WUE8 A0A0N0PAA2 A0A3S2NQU9 N6UPP4 A0A146KNU3 A0A087ZVE8 A0A151I9X6 A0A195CR36 A0A1W6BRF0 A0A224XNL2 A0A2D3FB68 A0A0V0G516 A0A1W5KJY8 A0A0T6BDP9 A0A0L7QRY7 A0A195DZH4 A0A2A3E8M7 A0A069DUW8 A0A2P8YCD2 A0A158NNQ4
PDB
4QLA
E-value=8.42331e-145,
Score=1317
Ontologies
GO
Topology
Subcellular location
Endoplasmic reticulum membrane
Microsome membrane
Microsome membrane
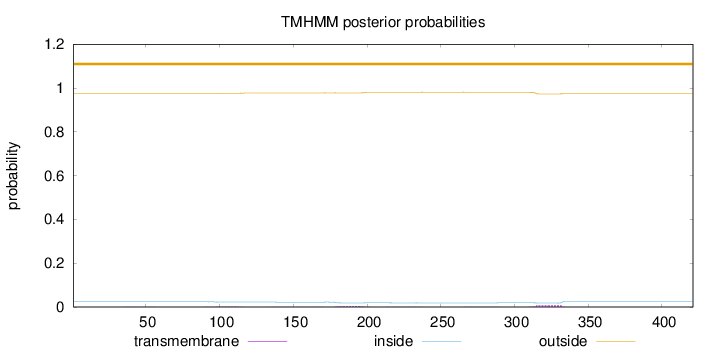
Length:
421
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.3126
Exp number, first 60 AAs:
0.00387
Total prob of N-in:
0.02405
outside
1 - 421
Population Genetic Test Statistics
Pi
250.860637
Theta
178.996374
Tajima's D
1.982858
CLR
0
CSRT
0.87995600219989
Interpretation
Uncertain
