Gene
KWMTBOMO16171
Pre Gene Modal
BGIBMGA013937
Annotation
PREDICTED:_lipoma_HMGIC_fusion_partner-like_3_protein_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.286 PlasmaMembrane Reliability : 1.75
Sequence
CDS
ATGCTTCGTCAAAAAGTACAATACATACTTTCAGCTGCAAGCGTCGTCATAACTCTGAGTCTGGTCGGCGGAGTGGCGGTGTACCCGGCCGGCTGGAACGAGACCTCAGTCCAGGAGACGTGCGGCCCCACAGCCGACCAATATAAACTAGGTGGCTGCCACATACGCTGGGCGTACATCTTGGCCGCGATAGGGTGTCTAGACGGCGCGGTGCTGTCTGCGCTCGCCTTCATCCTGGCCACGCGGCACGTGCGGCTGCGACCCGACGCCGGGTACCCGCCGCCGTCGCTATACAAAGGTGAGGTGAACAACGGCTACGTGACGGACGCGACATCTGTTAGCGGCTCTAGGAAGTCTCTGGCGCTGCAGCCCGTGCTCATCATGCACCCGCACGCGCCCATGGACATGGACACGTACTCTCACTACTCAGGACGCACCGCGCGATCCAAACACGGAATGTACGCGAACACTATGCATAATTATCAATTATAA
Protein
MLRQKVQYILSAASVVITLSLVGGVAVYPAGWNETSVQETCGPTADQYKLGGCHIRWAYILAAIGCLDGAVLSALAFILATRHVRLRPDAGYPPPSLYKGEVNNGYVTDATSVSGSRKSLALQPVLIMHPHAPMDMDTYSHYSGRTARSKHGMYANTMHNYQL
Summary
Uniprot
H9JWM3
A0A2H1WLU3
A0A2W1BAB6
A0A437B419
A0A2A4K8V8
S4PY30
+ More
A0A0N1IQD3 A0A194PZ34 I4DR84 I4DJM9 A0A212EWM5 A0A0L7LKZ8 A0A2J7Q9H2 A0A0V0G8J7 A0A0P4VPE7 A0A069DYH0 A0A1Y1LS63 A0A067RJL6 D6WPM0 A0A2R7X6U8 A0A1B6C8B7 A0A1B6LDF8 A0A1B0FJ69 A0A1A9ZMT0 A0A0A9XPG6 A0A1A9W4L8 A0A146MCC5 A0A1A9UXU7 A0A1B6JEZ0 A0A0L0CKE4 A0A1A9XBJ4 A0A1B0BAC6 A0A1I8N0P7 J3JYY7 A0A023F344 W8BXG3 A0A0A1WNB7 A0A2P8Z492 A0A034VTF9 A0A1B0GM04 A0A0K8TZ73 A0A2H8U0L5 A0A1B0CP82 A0A1S3D269 J9JJ24 A0A1L8D9H4 A0A2S2PNW7 A0A1L8D9E4 A0A1W4XQZ3 A0A336M2F8 A0A182JKB5 B4MXH2 A0A0M3QW20 E0VQQ3 B4GS36 B4KWW5 B4IWQ3 B4LH07 B3M784 A0A3B0JUZ4 Q2M056 A0A182FE58 A0A1W4VVH9 A0A2R5LM32 B4PE89 B3NEX5 Q178A0 A0A023EMD6 Q9W068 B4HW25 B4QN04 B0X8Z6 B7P3U6 W5JTW0 A0A087TVG2 A0A084WDL8 A0A2P6KRE9 A0A443SPZ7 A0A2M3ZBD3 A0A2M4BX53 A0A2M4CSD7 A0A2M4AT92 A0A087U3B4 A0A1B6HK69 U5ETD1 A0A0K8VVW8 A0A0A1WNT4 W8BLY2 A0A034VW39 A0A0K8VGI5 A0A443RBI5 A0A0K2U772
A0A0N1IQD3 A0A194PZ34 I4DR84 I4DJM9 A0A212EWM5 A0A0L7LKZ8 A0A2J7Q9H2 A0A0V0G8J7 A0A0P4VPE7 A0A069DYH0 A0A1Y1LS63 A0A067RJL6 D6WPM0 A0A2R7X6U8 A0A1B6C8B7 A0A1B6LDF8 A0A1B0FJ69 A0A1A9ZMT0 A0A0A9XPG6 A0A1A9W4L8 A0A146MCC5 A0A1A9UXU7 A0A1B6JEZ0 A0A0L0CKE4 A0A1A9XBJ4 A0A1B0BAC6 A0A1I8N0P7 J3JYY7 A0A023F344 W8BXG3 A0A0A1WNB7 A0A2P8Z492 A0A034VTF9 A0A1B0GM04 A0A0K8TZ73 A0A2H8U0L5 A0A1B0CP82 A0A1S3D269 J9JJ24 A0A1L8D9H4 A0A2S2PNW7 A0A1L8D9E4 A0A1W4XQZ3 A0A336M2F8 A0A182JKB5 B4MXH2 A0A0M3QW20 E0VQQ3 B4GS36 B4KWW5 B4IWQ3 B4LH07 B3M784 A0A3B0JUZ4 Q2M056 A0A182FE58 A0A1W4VVH9 A0A2R5LM32 B4PE89 B3NEX5 Q178A0 A0A023EMD6 Q9W068 B4HW25 B4QN04 B0X8Z6 B7P3U6 W5JTW0 A0A087TVG2 A0A084WDL8 A0A2P6KRE9 A0A443SPZ7 A0A2M3ZBD3 A0A2M4BX53 A0A2M4CSD7 A0A2M4AT92 A0A087U3B4 A0A1B6HK69 U5ETD1 A0A0K8VVW8 A0A0A1WNT4 W8BLY2 A0A034VW39 A0A0K8VGI5 A0A443RBI5 A0A0K2U772
Pubmed
19121390
28756777
23622113
26354079
22651552
22118469
+ More
26227816 27129103 26334808 28004739 24845553 18362917 19820115 25401762 26823975 26108605 25315136 22516182 23537049 25474469 24495485 25830018 29403074 25348373 17994087 20566863 18057021 15632085 17550304 17510324 24945155 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 20920257 23761445 24438588
26227816 27129103 26334808 28004739 24845553 18362917 19820115 25401762 26823975 26108605 25315136 22516182 23537049 25474469 24495485 25830018 29403074 25348373 17994087 20566863 18057021 15632085 17550304 17510324 24945155 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 20920257 23761445 24438588
EMBL
BABH01018358
BABH01018359
ODYU01009507
SOQ53998.1
KZ150184
PZC72469.1
+ More
RSAL01000177 RVE45014.1 NWSH01000031 PCG80509.1 GAIX01004003 JAA88557.1 KQ459764 KPJ20102.1 KQ459583 KPI98592.1 AK404991 BAM20424.1 AK401497 BAM18119.1 AGBW02011937 OWR45886.1 JTDY01000784 KOB75886.1 NEVH01016358 PNF25219.1 GECL01001768 JAP04356.1 GDKW01002372 JAI54223.1 GBGD01002360 JAC86529.1 GEZM01051739 JAV74725.1 KK852471 KDR23178.1 KQ971354 EFA06835.1 KK857844 PTY27509.1 GEDC01027562 GEDC01015580 JAS09736.1 JAS21718.1 GEBQ01018242 JAT21735.1 CCAG010018370 CCAG010018371 GBHO01021012 JAG22592.1 GDHC01001201 GDHC01000494 JAQ17428.1 JAQ18135.1 GECU01009987 JAS97719.1 JRES01000281 KNC32731.1 JXJN01010919 APGK01054057 APGK01054058 BT128468 KB741247 KB632281 AEE63425.1 ENN71981.1 ERL91317.1 GBBI01002912 JAC15800.1 GAMC01008559 JAB97996.1 GBXI01013995 JAD00297.1 PYGN01000203 PSN51312.1 GAKP01013243 JAC45709.1 AJVK01020176 GDHF01032731 GDHF01005569 JAI19583.1 JAI46745.1 GFXV01007143 MBW18948.1 AJWK01021534 ABLF02020856 GFDF01011130 JAV02954.1 GGMR01018542 MBY31161.1 GFDF01011079 JAV03005.1 UFQS01000305 UFQT01000305 SSX02689.1 SSX23063.1 CH963876 EDW76741.1 CP012525 ALC43383.1 DS235430 EEB15709.1 CH479188 EDW40571.1 CH933809 EDW18586.1 CH916366 EDV96279.1 CH940647 EDW69497.1 KRF84421.1 CH902618 EDV38745.1 OUUW01000009 SPP84883.1 CH379069 EAL31080.1 GGLE01006423 MBY10549.1 CM000159 EDW92927.1 CH954178 EDV50248.1 KQS43130.1 CH477368 EAT42484.1 JXUM01011305 JXUM01011306 JXUM01011307 JXUM01011308 JXUM01011309 JXUM01011310 GAPW01003126 KQ560328 JAC10472.1 KXJ83015.1 AE014296 AY052063 AAF47590.1 AAK93487.1 AAN11506.1 CH480817 EDW50140.1 CM000363 CM002912 EDX08886.1 KMY96984.1 DS232512 EDS42903.1 ABJB010638926 DS630485 EEC01268.1 ADMH02000499 ETN66189.1 KK116926 KFM69101.1 ATLV01023045 KE525339 KFB48312.1 MWRG01006528 PRD28899.1 NCKV01000845 RWS29594.1 GGFM01005071 MBW25822.1 GGFJ01008167 MBW57308.1 GGFL01004076 MBW68254.1 GGFK01010663 MBW43984.1 KK117954 KFM71853.1 GECU01032731 JAS74975.1 GANO01002835 JAB57036.1 GDHF01009295 JAI43019.1 GBXI01013996 JAD00296.1 GAMC01008557 JAB97998.1 GAKP01013244 JAC45708.1 GDHF01014351 JAI37963.1 NCKU01001244 RWS12631.1 HACA01016727 HACA01016729 CDW34088.1
RSAL01000177 RVE45014.1 NWSH01000031 PCG80509.1 GAIX01004003 JAA88557.1 KQ459764 KPJ20102.1 KQ459583 KPI98592.1 AK404991 BAM20424.1 AK401497 BAM18119.1 AGBW02011937 OWR45886.1 JTDY01000784 KOB75886.1 NEVH01016358 PNF25219.1 GECL01001768 JAP04356.1 GDKW01002372 JAI54223.1 GBGD01002360 JAC86529.1 GEZM01051739 JAV74725.1 KK852471 KDR23178.1 KQ971354 EFA06835.1 KK857844 PTY27509.1 GEDC01027562 GEDC01015580 JAS09736.1 JAS21718.1 GEBQ01018242 JAT21735.1 CCAG010018370 CCAG010018371 GBHO01021012 JAG22592.1 GDHC01001201 GDHC01000494 JAQ17428.1 JAQ18135.1 GECU01009987 JAS97719.1 JRES01000281 KNC32731.1 JXJN01010919 APGK01054057 APGK01054058 BT128468 KB741247 KB632281 AEE63425.1 ENN71981.1 ERL91317.1 GBBI01002912 JAC15800.1 GAMC01008559 JAB97996.1 GBXI01013995 JAD00297.1 PYGN01000203 PSN51312.1 GAKP01013243 JAC45709.1 AJVK01020176 GDHF01032731 GDHF01005569 JAI19583.1 JAI46745.1 GFXV01007143 MBW18948.1 AJWK01021534 ABLF02020856 GFDF01011130 JAV02954.1 GGMR01018542 MBY31161.1 GFDF01011079 JAV03005.1 UFQS01000305 UFQT01000305 SSX02689.1 SSX23063.1 CH963876 EDW76741.1 CP012525 ALC43383.1 DS235430 EEB15709.1 CH479188 EDW40571.1 CH933809 EDW18586.1 CH916366 EDV96279.1 CH940647 EDW69497.1 KRF84421.1 CH902618 EDV38745.1 OUUW01000009 SPP84883.1 CH379069 EAL31080.1 GGLE01006423 MBY10549.1 CM000159 EDW92927.1 CH954178 EDV50248.1 KQS43130.1 CH477368 EAT42484.1 JXUM01011305 JXUM01011306 JXUM01011307 JXUM01011308 JXUM01011309 JXUM01011310 GAPW01003126 KQ560328 JAC10472.1 KXJ83015.1 AE014296 AY052063 AAF47590.1 AAK93487.1 AAN11506.1 CH480817 EDW50140.1 CM000363 CM002912 EDX08886.1 KMY96984.1 DS232512 EDS42903.1 ABJB010638926 DS630485 EEC01268.1 ADMH02000499 ETN66189.1 KK116926 KFM69101.1 ATLV01023045 KE525339 KFB48312.1 MWRG01006528 PRD28899.1 NCKV01000845 RWS29594.1 GGFM01005071 MBW25822.1 GGFJ01008167 MBW57308.1 GGFL01004076 MBW68254.1 GGFK01010663 MBW43984.1 KK117954 KFM71853.1 GECU01032731 JAS74975.1 GANO01002835 JAB57036.1 GDHF01009295 JAI43019.1 GBXI01013996 JAD00296.1 GAMC01008557 JAB97998.1 GAKP01013244 JAC45708.1 GDHF01014351 JAI37963.1 NCKU01001244 RWS12631.1 HACA01016727 HACA01016729 CDW34088.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000037510 UP000235965 UP000027135 UP000007266 UP000092444 UP000092445 UP000091820 UP000078200 UP000037069 UP000092443 UP000092460 UP000095301 UP000019118 UP000030742 UP000245037 UP000092462 UP000092461 UP000079169 UP000007819 UP000192223 UP000075880 UP000007798 UP000092553 UP000009046 UP000008744 UP000009192 UP000001070 UP000008792 UP000007801 UP000268350 UP000001819 UP000069272 UP000192221 UP000002282 UP000008711 UP000008820 UP000069940 UP000249989 UP000000803 UP000001292 UP000000304 UP000002320 UP000001555 UP000000673 UP000054359 UP000030765 UP000288716 UP000285301
UP000037510 UP000235965 UP000027135 UP000007266 UP000092444 UP000092445 UP000091820 UP000078200 UP000037069 UP000092443 UP000092460 UP000095301 UP000019118 UP000030742 UP000245037 UP000092462 UP000092461 UP000079169 UP000007819 UP000192223 UP000075880 UP000007798 UP000092553 UP000009046 UP000008744 UP000009192 UP000001070 UP000008792 UP000007801 UP000268350 UP000001819 UP000069272 UP000192221 UP000002282 UP000008711 UP000008820 UP000069940 UP000249989 UP000000803 UP000001292 UP000000304 UP000002320 UP000001555 UP000000673 UP000054359 UP000030765 UP000288716 UP000285301
Interpro
SUPFAM
SSF53271
SSF53271
ProteinModelPortal
H9JWM3
A0A2H1WLU3
A0A2W1BAB6
A0A437B419
A0A2A4K8V8
S4PY30
+ More
A0A0N1IQD3 A0A194PZ34 I4DR84 I4DJM9 A0A212EWM5 A0A0L7LKZ8 A0A2J7Q9H2 A0A0V0G8J7 A0A0P4VPE7 A0A069DYH0 A0A1Y1LS63 A0A067RJL6 D6WPM0 A0A2R7X6U8 A0A1B6C8B7 A0A1B6LDF8 A0A1B0FJ69 A0A1A9ZMT0 A0A0A9XPG6 A0A1A9W4L8 A0A146MCC5 A0A1A9UXU7 A0A1B6JEZ0 A0A0L0CKE4 A0A1A9XBJ4 A0A1B0BAC6 A0A1I8N0P7 J3JYY7 A0A023F344 W8BXG3 A0A0A1WNB7 A0A2P8Z492 A0A034VTF9 A0A1B0GM04 A0A0K8TZ73 A0A2H8U0L5 A0A1B0CP82 A0A1S3D269 J9JJ24 A0A1L8D9H4 A0A2S2PNW7 A0A1L8D9E4 A0A1W4XQZ3 A0A336M2F8 A0A182JKB5 B4MXH2 A0A0M3QW20 E0VQQ3 B4GS36 B4KWW5 B4IWQ3 B4LH07 B3M784 A0A3B0JUZ4 Q2M056 A0A182FE58 A0A1W4VVH9 A0A2R5LM32 B4PE89 B3NEX5 Q178A0 A0A023EMD6 Q9W068 B4HW25 B4QN04 B0X8Z6 B7P3U6 W5JTW0 A0A087TVG2 A0A084WDL8 A0A2P6KRE9 A0A443SPZ7 A0A2M3ZBD3 A0A2M4BX53 A0A2M4CSD7 A0A2M4AT92 A0A087U3B4 A0A1B6HK69 U5ETD1 A0A0K8VVW8 A0A0A1WNT4 W8BLY2 A0A034VW39 A0A0K8VGI5 A0A443RBI5 A0A0K2U772
A0A0N1IQD3 A0A194PZ34 I4DR84 I4DJM9 A0A212EWM5 A0A0L7LKZ8 A0A2J7Q9H2 A0A0V0G8J7 A0A0P4VPE7 A0A069DYH0 A0A1Y1LS63 A0A067RJL6 D6WPM0 A0A2R7X6U8 A0A1B6C8B7 A0A1B6LDF8 A0A1B0FJ69 A0A1A9ZMT0 A0A0A9XPG6 A0A1A9W4L8 A0A146MCC5 A0A1A9UXU7 A0A1B6JEZ0 A0A0L0CKE4 A0A1A9XBJ4 A0A1B0BAC6 A0A1I8N0P7 J3JYY7 A0A023F344 W8BXG3 A0A0A1WNB7 A0A2P8Z492 A0A034VTF9 A0A1B0GM04 A0A0K8TZ73 A0A2H8U0L5 A0A1B0CP82 A0A1S3D269 J9JJ24 A0A1L8D9H4 A0A2S2PNW7 A0A1L8D9E4 A0A1W4XQZ3 A0A336M2F8 A0A182JKB5 B4MXH2 A0A0M3QW20 E0VQQ3 B4GS36 B4KWW5 B4IWQ3 B4LH07 B3M784 A0A3B0JUZ4 Q2M056 A0A182FE58 A0A1W4VVH9 A0A2R5LM32 B4PE89 B3NEX5 Q178A0 A0A023EMD6 Q9W068 B4HW25 B4QN04 B0X8Z6 B7P3U6 W5JTW0 A0A087TVG2 A0A084WDL8 A0A2P6KRE9 A0A443SPZ7 A0A2M3ZBD3 A0A2M4BX53 A0A2M4CSD7 A0A2M4AT92 A0A087U3B4 A0A1B6HK69 U5ETD1 A0A0K8VVW8 A0A0A1WNT4 W8BLY2 A0A034VW39 A0A0K8VGI5 A0A443RBI5 A0A0K2U772
PDB
6C14
E-value=3.21866e-12,
Score=168
Ontologies
PANTHER
Topology
SignalP
Position: 1 - 24,
Likelihood: 0.735640
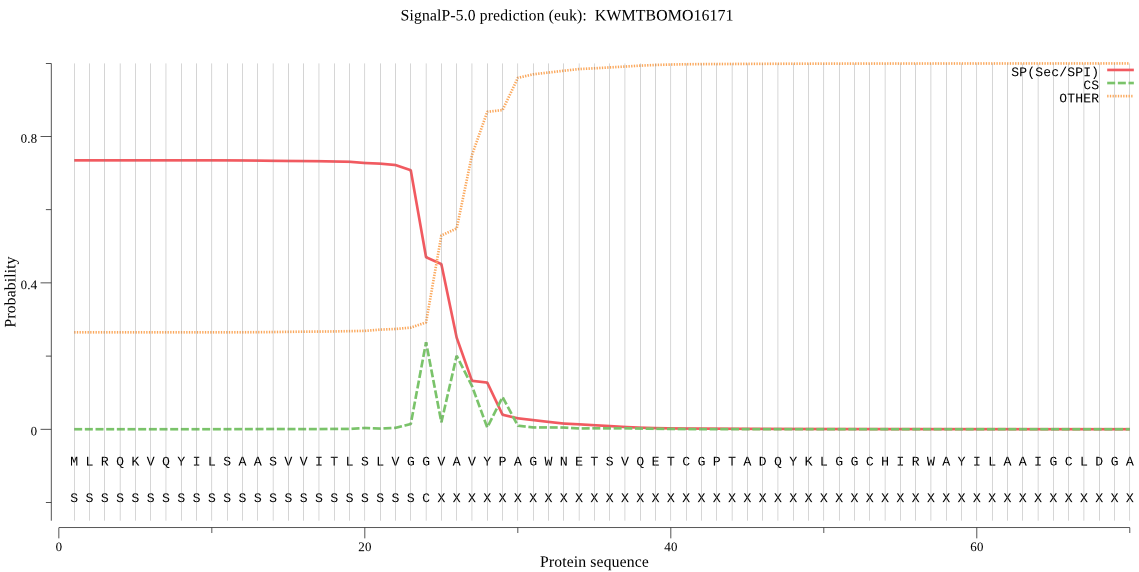
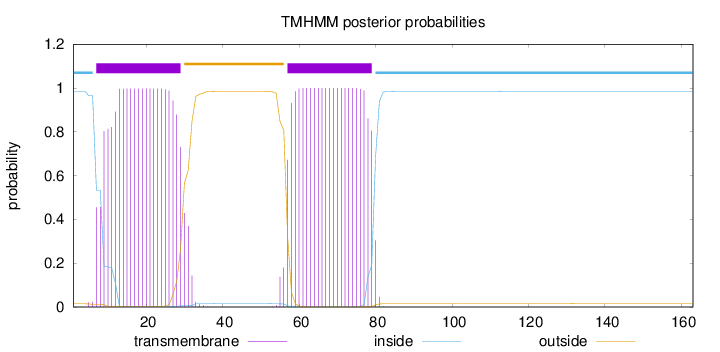
Length:
163
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
44.6943400000001
Exp number, first 60 AAs:
25.68962
Total prob of N-in:
0.98545
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 56
TMhelix
57 - 79
inside
80 - 163
Population Genetic Test Statistics
Pi
205.68963
Theta
55.036001
Tajima's D
1.140973
CLR
0.064231
CSRT
0.701364931753412
Interpretation
Uncertain
