Gene
KWMTBOMO16170
Pre Gene Modal
BGIBMGA013955
Annotation
PREDICTED:_protein_N-lysine_methyltransferase_METTL20-like_isoform_X3_[Bombyx_mori]
Full name
Electron transfer flavoprotein beta subunit lysine methyltransferase
Alternative Name
ETFB lysine methyltransferase
Protein N-lysine methyltransferase METTL20
Protein N-lysine methyltransferase METTL20
Location in the cell
Mitochondrial Reliability : 2.031
Sequence
CDS
ATGTTGAGAGACATTTCGGCTGCCATCATAAGGCACACGGCCTTATCGCGACGACACCTGACTCCCGAGCTGGTTCTGCGTCTCATCACACCGGAATGTCCGCTATGGAGAGCTAAAGAAGGCGAGTCGCCTTTCAGAGACCCGTTCTGGGCATTCTATTGGCCCGGAGGTCAGGCTACTGCGCGGTACATACTAGACAACCAAGACTTGGTAAAAGATCGTAATGTATTAGACATTGGATGTGGTTGCGGAGCGGGATCAATTGCGGCCGCTAAAATGAAGGCAAAATATGTTTTGGCCAACGACATTGACCACCATGCACTTGCAGCAACACATCTGAATGCAAATTTAAATGGAGTACTAATTGATACTAGTTCCAAGAATTTAATTGGTACAAAATGTGAAGAGTTCAATGTGATTTTGATCGGTGATATGTTTTACGATGAAGATTTTTCTCAGACACTATTCAAATGGCTAATTAAATTGCACGAAGCAGGAAAAACGATACTAATAGGAGATCCAGGTCGTCACGGCTTGACAGAGAAGCGTAGACGACATGTGCAGATGCTCGCCGAGTACAAGATGCCCAGGGAGAGCCAAGAGGAGAACAACGGTTTTACGAAGACTAAAGTTTGGAAGTTAAATAAATTGATTTGA
Protein
MLRDISAAIIRHTALSRRHLTPELVLRLITPECPLWRAKEGESPFRDPFWAFYWPGGQATARYILDNQDLVKDRNVLDIGCGCGAGSIAAAKMKAKYVLANDIDHHALAATHLNANLNGVLIDTSSKNLIGTKCEEFNVILIGDMFYDEDFSQTLFKWLIKLHEAGKTILIGDPGRHGLTEKRRRHVQMLAEYKMPRESQEENNGFTKTKVWKLNKLI
Summary
Description
Protein-lysine methyltransferase that selectively trimethylates the flavoprotein ETFB in mitochondria. Thereby, may negatively regulate the function of ETFB in electron transfer from Acyl-CoA dehydrogenases to the main respiratory chain.
Subunit
Interacts with HSPD1; this protein may possibly be a methylation substrate.
Similarity
Belongs to the MT-A70-like family.
Belongs to the methyltransferase superfamily. ETFBKMT family.
Belongs to the methyltransferase superfamily. ETFBKMT family.
Keywords
Complete proteome
Cytoplasm
Methyltransferase
Mitochondrion
Reference proteome
Transferase
Transit peptide
Feature
chain Electron transfer flavoprotein beta subunit lysine methyltransferase
Uniprot
A0A437B4I6
H9JWP1
A0A2W1BLC3
A0A1E1WAJ1
A0A0N1IJW4
A0A2P8Y016
+ More
A0A194PZ45 R7VE04 A0A034VYL6 A0A210Q6L2 A0A0K8WIP6 A0A0L0CAM2 A0A1A9W128 A0A0A1XJP6 E0VWI7 A0A212EWR1 A0A1B0AAS2 A0A0A9W7Z3 A0A1B6BY50 W8CDF1 A0A1B0FP85 A0A1I8MQ77 J9K2C9 X1WI67 A0A1I8PSM1 A0A146LS71 V4A0W6 A0A0B6YS33 A0A2M4CWH8 A0A2M4AXI1 A0A0P4WHH7 A0A2A4K9D0 A0A0P5F1F7 A0A2M4CWI3 A0A444U4S1 A0A0P5Z2B4 A0A433TP12 E9HBZ1 A0A182IY09 Q174H9 K1R819 K7FXQ9 A0A0P5FUA0 Q4SM76 C3Z9A5 A0A3B3TF80 A0A1S3HVU2 A0A401REG6 A0A182P9W0 A0A182FIB1 A0A2M4CX05 H0ZMK5 A0A402EM00 A0A091GRC4 A0A094KVT4 K7IX40 A0A1S3PCL5 A0A091KBW8 A0A3Q1CUX6 A0A060WDU7 R4GLZ9 A0A091NGY5 H3AZ38 A0A091I1B0 A0A093CLW1 A0A093PAE8 A0A091UXQ8 A0A1J1ISQ6 A0A093IJ64 A0A091LPR0 Q6P7Q0 A0A0P6C3C0 A0A091VL41 A0A091QZM9 A0A0A0ADA5 A0A3Q0RKY2 A0A151NIS3 A0A091SCX9 A0A3P8SX37 A0A3B3YL61 A0A093FRM0 I3JWA6 A0A3B4VBZ3 A0A091KNQ8 I3JWA5 A0A2M4CWJ0 A0A091PBR0 A0A3B4XBU7 A0A1U7QRA2 A0A1B6FX26 A0A226MT24 A0A182TG15 A0A093QBZ2 A0A099ZNE8 A0A087RJF6 A0A182K3M4 A0A154PRG4 A0A087V914 A0A091SY32 A0A3Q3JHB1
A0A194PZ45 R7VE04 A0A034VYL6 A0A210Q6L2 A0A0K8WIP6 A0A0L0CAM2 A0A1A9W128 A0A0A1XJP6 E0VWI7 A0A212EWR1 A0A1B0AAS2 A0A0A9W7Z3 A0A1B6BY50 W8CDF1 A0A1B0FP85 A0A1I8MQ77 J9K2C9 X1WI67 A0A1I8PSM1 A0A146LS71 V4A0W6 A0A0B6YS33 A0A2M4CWH8 A0A2M4AXI1 A0A0P4WHH7 A0A2A4K9D0 A0A0P5F1F7 A0A2M4CWI3 A0A444U4S1 A0A0P5Z2B4 A0A433TP12 E9HBZ1 A0A182IY09 Q174H9 K1R819 K7FXQ9 A0A0P5FUA0 Q4SM76 C3Z9A5 A0A3B3TF80 A0A1S3HVU2 A0A401REG6 A0A182P9W0 A0A182FIB1 A0A2M4CX05 H0ZMK5 A0A402EM00 A0A091GRC4 A0A094KVT4 K7IX40 A0A1S3PCL5 A0A091KBW8 A0A3Q1CUX6 A0A060WDU7 R4GLZ9 A0A091NGY5 H3AZ38 A0A091I1B0 A0A093CLW1 A0A093PAE8 A0A091UXQ8 A0A1J1ISQ6 A0A093IJ64 A0A091LPR0 Q6P7Q0 A0A0P6C3C0 A0A091VL41 A0A091QZM9 A0A0A0ADA5 A0A3Q0RKY2 A0A151NIS3 A0A091SCX9 A0A3P8SX37 A0A3B3YL61 A0A093FRM0 I3JWA6 A0A3B4VBZ3 A0A091KNQ8 I3JWA5 A0A2M4CWJ0 A0A091PBR0 A0A3B4XBU7 A0A1U7QRA2 A0A1B6FX26 A0A226MT24 A0A182TG15 A0A093QBZ2 A0A099ZNE8 A0A087RJF6 A0A182K3M4 A0A154PRG4 A0A087V914 A0A091SY32 A0A3Q3JHB1
EC Number
2.1.1.-
Pubmed
EMBL
RSAL01000177
RVE45015.1
BABH01018358
KZ150184
PZC72473.1
GDQN01007089
+ More
JAT83965.1 KQ459764 KPJ20103.1 PYGN01001100 PSN37598.1 KQ459583 KPI98591.1 AMQN01004815 KB294418 ELU14536.1 GAKP01011982 JAC46970.1 NEDP02004827 OWF44329.1 GDHF01001286 JAI51028.1 JRES01000678 KNC29301.1 GBXI01009301 GBXI01002703 JAD04991.1 JAD11589.1 DS235821 EEB17743.1 AGBW02011937 OWR45887.1 GBHO01039047 JAG04557.1 GEDC01031086 JAS06212.1 GAMC01002056 JAC04500.1 CCAG010011789 ABLF02024323 ABLF02024318 GDHC01007981 JAQ10648.1 KB201305 ESO97448.1 HACG01012148 HACG01012149 CEK59013.1 CEK59014.1 GGFL01005515 MBW69693.1 GGFK01012134 MBW45455.1 GDRN01052876 GDRN01052875 GDRN01052874 JAI66234.1 NWSH01000031 PCG80498.1 GDIQ01269004 JAJ82720.1 GGFL01005516 MBW69694.1 SCEB01215313 RXM30164.1 GDIP01051564 LRGB01000359 JAM52151.1 KZS19581.1 RQTK01000249 RUS83314.1 GL732618 EFX70673.1 CH477410 EAT41479.1 JH818716 EKC37350.1 AGCU01028385 AGCU01028386 GDIQ01249957 JAK01768.1 CAAE01014555 CAF98256.1 GG666599 EEN50834.1 BEZZ01003823 GCC16534.1 GGFL01005672 MBW69850.1 ABQF01016311 BDOT01000023 GCF45168.1 KL506511 KFO85027.1 KL345858 KFZ55720.1 AAZX01000615 AAZX01009233 KK553661 KFP34525.1 FR904508 CDQ65478.1 AADN05000523 KL383496 KFP89028.1 AFYH01126962 KL218095 KFP02309.1 KL244638 KFV13939.1 KL225530 KFW73401.1 KK466903 KFQ82383.1 CVRI01000059 CRL03231.1 KL215805 KFV66756.1 KL334465 KFP59148.1 BC061574 GDIP01009933 JAM93782.1 KK436010 KFQ90258.1 KK806044 KFQ32141.1 KL871447 KGL92131.1 AKHW03002956 KYO36559.1 KK948302 KFQ56173.1 KK401838 KFV60417.1 AERX01017318 AERX01017319 KK748133 KFP40920.1 GGFL01005518 MBW69696.1 KK655459 KFQ04628.1 GECZ01015045 JAS54724.1 MCFN01000467 OXB58447.1 KL671842 KFW83950.1 KL895188 KGL82443.1 KL226405 KFM13610.1 KQ435078 KZC14506.1 KL484389 KFO09106.1 KK491519 KFQ63642.1
JAT83965.1 KQ459764 KPJ20103.1 PYGN01001100 PSN37598.1 KQ459583 KPI98591.1 AMQN01004815 KB294418 ELU14536.1 GAKP01011982 JAC46970.1 NEDP02004827 OWF44329.1 GDHF01001286 JAI51028.1 JRES01000678 KNC29301.1 GBXI01009301 GBXI01002703 JAD04991.1 JAD11589.1 DS235821 EEB17743.1 AGBW02011937 OWR45887.1 GBHO01039047 JAG04557.1 GEDC01031086 JAS06212.1 GAMC01002056 JAC04500.1 CCAG010011789 ABLF02024323 ABLF02024318 GDHC01007981 JAQ10648.1 KB201305 ESO97448.1 HACG01012148 HACG01012149 CEK59013.1 CEK59014.1 GGFL01005515 MBW69693.1 GGFK01012134 MBW45455.1 GDRN01052876 GDRN01052875 GDRN01052874 JAI66234.1 NWSH01000031 PCG80498.1 GDIQ01269004 JAJ82720.1 GGFL01005516 MBW69694.1 SCEB01215313 RXM30164.1 GDIP01051564 LRGB01000359 JAM52151.1 KZS19581.1 RQTK01000249 RUS83314.1 GL732618 EFX70673.1 CH477410 EAT41479.1 JH818716 EKC37350.1 AGCU01028385 AGCU01028386 GDIQ01249957 JAK01768.1 CAAE01014555 CAF98256.1 GG666599 EEN50834.1 BEZZ01003823 GCC16534.1 GGFL01005672 MBW69850.1 ABQF01016311 BDOT01000023 GCF45168.1 KL506511 KFO85027.1 KL345858 KFZ55720.1 AAZX01000615 AAZX01009233 KK553661 KFP34525.1 FR904508 CDQ65478.1 AADN05000523 KL383496 KFP89028.1 AFYH01126962 KL218095 KFP02309.1 KL244638 KFV13939.1 KL225530 KFW73401.1 KK466903 KFQ82383.1 CVRI01000059 CRL03231.1 KL215805 KFV66756.1 KL334465 KFP59148.1 BC061574 GDIP01009933 JAM93782.1 KK436010 KFQ90258.1 KK806044 KFQ32141.1 KL871447 KGL92131.1 AKHW03002956 KYO36559.1 KK948302 KFQ56173.1 KK401838 KFV60417.1 AERX01017318 AERX01017319 KK748133 KFP40920.1 GGFL01005518 MBW69696.1 KK655459 KFQ04628.1 GECZ01015045 JAS54724.1 MCFN01000467 OXB58447.1 KL671842 KFW83950.1 KL895188 KGL82443.1 KL226405 KFM13610.1 KQ435078 KZC14506.1 KL484389 KFO09106.1 KK491519 KFQ63642.1
Proteomes
UP000283053
UP000005204
UP000053240
UP000245037
UP000053268
UP000014760
+ More
UP000242188 UP000037069 UP000091820 UP000009046 UP000007151 UP000092445 UP000092444 UP000095301 UP000007819 UP000095300 UP000030746 UP000218220 UP000076858 UP000271974 UP000000305 UP000075880 UP000008820 UP000005408 UP000007267 UP000007303 UP000001554 UP000261540 UP000085678 UP000287033 UP000075885 UP000069272 UP000007754 UP000288954 UP000002358 UP000087266 UP000257160 UP000193380 UP000000539 UP000008672 UP000054308 UP000054081 UP000183832 UP000053875 UP000002494 UP000053858 UP000261340 UP000050525 UP000265080 UP000261480 UP000005207 UP000261420 UP000261360 UP000189706 UP000198323 UP000075902 UP000053258 UP000053641 UP000053286 UP000075881 UP000076502 UP000261600
UP000242188 UP000037069 UP000091820 UP000009046 UP000007151 UP000092445 UP000092444 UP000095301 UP000007819 UP000095300 UP000030746 UP000218220 UP000076858 UP000271974 UP000000305 UP000075880 UP000008820 UP000005408 UP000007267 UP000007303 UP000001554 UP000261540 UP000085678 UP000287033 UP000075885 UP000069272 UP000007754 UP000288954 UP000002358 UP000087266 UP000257160 UP000193380 UP000000539 UP000008672 UP000054308 UP000054081 UP000183832 UP000053875 UP000002494 UP000053858 UP000261340 UP000050525 UP000265080 UP000261480 UP000005207 UP000261420 UP000261360 UP000189706 UP000198323 UP000075902 UP000053258 UP000053641 UP000053286 UP000075881 UP000076502 UP000261600
Interpro
SUPFAM
SSF53335
SSF53335
Gene 3D
ProteinModelPortal
A0A437B4I6
H9JWP1
A0A2W1BLC3
A0A1E1WAJ1
A0A0N1IJW4
A0A2P8Y016
+ More
A0A194PZ45 R7VE04 A0A034VYL6 A0A210Q6L2 A0A0K8WIP6 A0A0L0CAM2 A0A1A9W128 A0A0A1XJP6 E0VWI7 A0A212EWR1 A0A1B0AAS2 A0A0A9W7Z3 A0A1B6BY50 W8CDF1 A0A1B0FP85 A0A1I8MQ77 J9K2C9 X1WI67 A0A1I8PSM1 A0A146LS71 V4A0W6 A0A0B6YS33 A0A2M4CWH8 A0A2M4AXI1 A0A0P4WHH7 A0A2A4K9D0 A0A0P5F1F7 A0A2M4CWI3 A0A444U4S1 A0A0P5Z2B4 A0A433TP12 E9HBZ1 A0A182IY09 Q174H9 K1R819 K7FXQ9 A0A0P5FUA0 Q4SM76 C3Z9A5 A0A3B3TF80 A0A1S3HVU2 A0A401REG6 A0A182P9W0 A0A182FIB1 A0A2M4CX05 H0ZMK5 A0A402EM00 A0A091GRC4 A0A094KVT4 K7IX40 A0A1S3PCL5 A0A091KBW8 A0A3Q1CUX6 A0A060WDU7 R4GLZ9 A0A091NGY5 H3AZ38 A0A091I1B0 A0A093CLW1 A0A093PAE8 A0A091UXQ8 A0A1J1ISQ6 A0A093IJ64 A0A091LPR0 Q6P7Q0 A0A0P6C3C0 A0A091VL41 A0A091QZM9 A0A0A0ADA5 A0A3Q0RKY2 A0A151NIS3 A0A091SCX9 A0A3P8SX37 A0A3B3YL61 A0A093FRM0 I3JWA6 A0A3B4VBZ3 A0A091KNQ8 I3JWA5 A0A2M4CWJ0 A0A091PBR0 A0A3B4XBU7 A0A1U7QRA2 A0A1B6FX26 A0A226MT24 A0A182TG15 A0A093QBZ2 A0A099ZNE8 A0A087RJF6 A0A182K3M4 A0A154PRG4 A0A087V914 A0A091SY32 A0A3Q3JHB1
A0A194PZ45 R7VE04 A0A034VYL6 A0A210Q6L2 A0A0K8WIP6 A0A0L0CAM2 A0A1A9W128 A0A0A1XJP6 E0VWI7 A0A212EWR1 A0A1B0AAS2 A0A0A9W7Z3 A0A1B6BY50 W8CDF1 A0A1B0FP85 A0A1I8MQ77 J9K2C9 X1WI67 A0A1I8PSM1 A0A146LS71 V4A0W6 A0A0B6YS33 A0A2M4CWH8 A0A2M4AXI1 A0A0P4WHH7 A0A2A4K9D0 A0A0P5F1F7 A0A2M4CWI3 A0A444U4S1 A0A0P5Z2B4 A0A433TP12 E9HBZ1 A0A182IY09 Q174H9 K1R819 K7FXQ9 A0A0P5FUA0 Q4SM76 C3Z9A5 A0A3B3TF80 A0A1S3HVU2 A0A401REG6 A0A182P9W0 A0A182FIB1 A0A2M4CX05 H0ZMK5 A0A402EM00 A0A091GRC4 A0A094KVT4 K7IX40 A0A1S3PCL5 A0A091KBW8 A0A3Q1CUX6 A0A060WDU7 R4GLZ9 A0A091NGY5 H3AZ38 A0A091I1B0 A0A093CLW1 A0A093PAE8 A0A091UXQ8 A0A1J1ISQ6 A0A093IJ64 A0A091LPR0 Q6P7Q0 A0A0P6C3C0 A0A091VL41 A0A091QZM9 A0A0A0ADA5 A0A3Q0RKY2 A0A151NIS3 A0A091SCX9 A0A3P8SX37 A0A3B3YL61 A0A093FRM0 I3JWA6 A0A3B4VBZ3 A0A091KNQ8 I3JWA5 A0A2M4CWJ0 A0A091PBR0 A0A3B4XBU7 A0A1U7QRA2 A0A1B6FX26 A0A226MT24 A0A182TG15 A0A093QBZ2 A0A099ZNE8 A0A087RJF6 A0A182K3M4 A0A154PRG4 A0A087V914 A0A091SY32 A0A3Q3JHB1
Ontologies
KEGG
GO
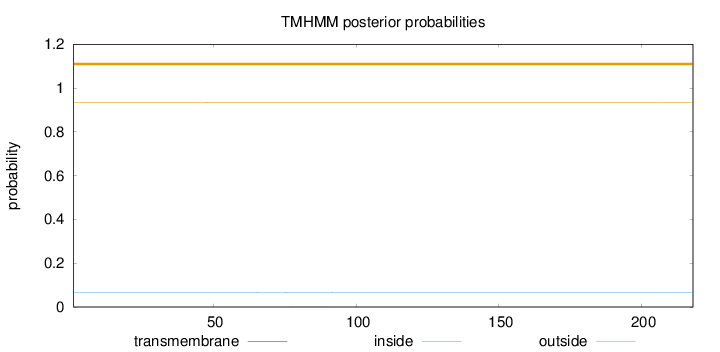
Topology
Subcellular location
Cytoplasm
Concentrated in cytoplasmic granular foci. With evidence from 2 publications.
Mitochondrion matrix Concentrated in cytoplasmic granular foci. With evidence from 2 publications.
Mitochondrion matrix Concentrated in cytoplasmic granular foci. With evidence from 2 publications.
Length:
218
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00528
Exp number, first 60 AAs:
0.00141
Total prob of N-in:
0.06656
outside
1 - 218
Population Genetic Test Statistics
Pi
28.085617
Theta
185.997012
Tajima's D
1.104203
CLR
0.360326
CSRT
0.690265486725664
Interpretation
Uncertain
