Gene
KWMTBOMO16169
Pre Gene Modal
BGIBMGA013938
Annotation
uncharacterized_LOC106115108_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 2.398
Sequence
CDS
ATGTACAAGGAAGCAGTTGTTCCAGTAAAAGCCGAGACCTTCAGTGTCATAGCAGCTAAAAGCTCTATCGGACAATGTATCAAGACAGTGTTAAGGCGAGCCTGTGTCGAAAAAAGGTTAACTATTGGCTTACTTCCCGCCATTCAATACTTATCGAAGAACTGCAATGGTGCATTGTTTTGCCTGACCGCCGAAGCCCCGCCTGGCGATAGTGCAACGCATATGCAAGATGTCCTCCTGCAAGCATTTTGCGTTGAAAATGATATTCATGTTATTAAAGTCGATTGTGAGACTAAGCTGAGGAGAATGCTTGGTTACTGCTCTCCCATGGACTTCAGTTGCGTGCTGGTACACTATCCGTATACAGATCCATTCACAGACAGTCAAGAAATAGATTTATCAACATTATCTGAAGCCGAGAGACAGTTGATCGACCACTGCGAATCAGACTGGGGCTATTCACAAATGCCCGTCATCAAGTTGCCCGAGAAGTGA
Protein
MYKEAVVPVKAETFSVIAAKSSIGQCIKTVLRRACVEKRLTIGLLPAIQYLSKNCNGALFCLTAEAPPGDSATHMQDVLLQAFCVENDIHVIKVDCETKLRRMLGYCSPMDFSCVLVHYPYTDPFTDSQEIDLSTLSEAERQLIDHCESDWGYSQMPVIKLPEK
Summary
Similarity
Belongs to the GMC oxidoreductase family.
Uniprot
H9JWM4
A0A2W1BFM2
A0A2H1X291
A0A437B3E2
I4DJN0
A0A194Q5C0
+ More
A0A0N1IIM5 A0A0L7KWX8 A0A212EW14 A0A212EW05 A0A0M4EC44 B4J411 B3MI60 A0A034VLZ1 B4LN81 A0A1I8P7R3 B4P1M3 A0A0A1XLZ6 A0A0J9R7A4 A1Z6W5 B3NAF6 Q8MYS8 B4KU01 B4HQS3 B4MPY9 A0A1W4VDR6 A0A1B0ALD5 A0A1I8NDQ8 A0A0K8VDL3 T1P8K8 A0A0L0CAK1 A0A1B0D0N2 A0A3B0J2A9 A0A1B0CGU0 A0A1A9W135 A0A1A9X9J2 Q292T6 W8BS98 A0A1B0AAS6 A0A1A9UDF0 B4GCN1 A0A1B0FPB9 A0A1L8DBJ1 A0A2P8Y037 A0A1Q3FLX6 B0WZR5 Q174I0 Q174I1 A0A2J7Q653 A0A1W7R5A5 A0A067RAM3 A0A1J1IU46 A0A182U9D5 A0A182V2D3 A0A2C9GRU2 Q7QJT5 A0A1J1ISN1 A0A182G162 A0A182P9V9 A0A182KRW6 A0A182LRN2 A0A182Q5S8 A0A1J1IXE1 A0A1J1ISX4 A0A336M5U4 A0A182XZH4 A0A182MD23 A0A182XZH3 A0A182RFQ5 A0A2M4C2W3 A0A084VF98 A0A2C9GRW5 A7UR75 A0A182VIG8 A0A182TQP5 A0A182X6G4 A0A1Y1KE78 A0A182Q0A0 A0A182KDS1 A0A182WIT8 A0A084VF99 A0A182KRW4 A0A1J1ISL1 A0A182JAT8 A0A2M4DS39 A0A0T6BCH8 A0A2M4AUQ1 A0A182NHP0 W5JTS6 A0A182IS99 D6WR71 A0A1B6M049 A0A182FIB2 A0A1B6F5Z9 A0A1B6H815 A0A2M4AUU6 A0A1W4X938 N6UHN0
A0A0N1IIM5 A0A0L7KWX8 A0A212EW14 A0A212EW05 A0A0M4EC44 B4J411 B3MI60 A0A034VLZ1 B4LN81 A0A1I8P7R3 B4P1M3 A0A0A1XLZ6 A0A0J9R7A4 A1Z6W5 B3NAF6 Q8MYS8 B4KU01 B4HQS3 B4MPY9 A0A1W4VDR6 A0A1B0ALD5 A0A1I8NDQ8 A0A0K8VDL3 T1P8K8 A0A0L0CAK1 A0A1B0D0N2 A0A3B0J2A9 A0A1B0CGU0 A0A1A9W135 A0A1A9X9J2 Q292T6 W8BS98 A0A1B0AAS6 A0A1A9UDF0 B4GCN1 A0A1B0FPB9 A0A1L8DBJ1 A0A2P8Y037 A0A1Q3FLX6 B0WZR5 Q174I0 Q174I1 A0A2J7Q653 A0A1W7R5A5 A0A067RAM3 A0A1J1IU46 A0A182U9D5 A0A182V2D3 A0A2C9GRU2 Q7QJT5 A0A1J1ISN1 A0A182G162 A0A182P9V9 A0A182KRW6 A0A182LRN2 A0A182Q5S8 A0A1J1IXE1 A0A1J1ISX4 A0A336M5U4 A0A182XZH4 A0A182MD23 A0A182XZH3 A0A182RFQ5 A0A2M4C2W3 A0A084VF98 A0A2C9GRW5 A7UR75 A0A182VIG8 A0A182TQP5 A0A182X6G4 A0A1Y1KE78 A0A182Q0A0 A0A182KDS1 A0A182WIT8 A0A084VF99 A0A182KRW4 A0A1J1ISL1 A0A182JAT8 A0A2M4DS39 A0A0T6BCH8 A0A2M4AUQ1 A0A182NHP0 W5JTS6 A0A182IS99 D6WR71 A0A1B6M049 A0A182FIB2 A0A1B6F5Z9 A0A1B6H815 A0A2M4AUU6 A0A1W4X938 N6UHN0
Pubmed
19121390
28756777
22651552
26354079
26227816
22118469
+ More
17994087 25348373 17550304 25830018 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 26108605 15632085 24495485 29403074 17510324 24845553 12364791 14747013 17210077 26483478 20966253 25244985 24438588 28004739 20920257 23761445 18362917 19820115 23537049
17994087 25348373 17550304 25830018 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 26108605 15632085 24495485 29403074 17510324 24845553 12364791 14747013 17210077 26483478 20966253 25244985 24438588 28004739 20920257 23761445 18362917 19820115 23537049
EMBL
BABH01018352
KZ150184
PZC72474.1
ODYU01012451
SOQ58764.1
RSAL01000177
+ More
RVE45017.1 AK401498 BAM18120.1 KQ459583 KPI98590.1 KQ459764 KPJ20104.1 JTDY01004797 KOB67742.1 AGBW02012100 OWR45673.1 OWR45672.1 CP012524 ALC41227.1 CH916367 EDW01594.1 CH902619 EDV35905.1 GAKP01014616 JAC44336.1 CH940648 EDW60085.1 CM000157 EDW89159.1 GBXI01002417 JAD11875.1 CM002911 KMY91936.1 AE013599 BT088861 AAF59278.1 ACS78061.1 CH954177 EDV59710.1 AY113626 AAM29631.1 CH933808 EDW08578.1 CH480816 EDW46733.1 CH963849 EDW74178.1 JXJN01030937 GDHF01021914 GDHF01015387 JAI30400.1 JAI36927.1 KA645096 AFP59725.1 JRES01000678 KNC29282.1 AJVK01010005 OUUW01000001 SPP73113.1 AJWK01011587 CM000071 EAL24775.1 GAMC01002325 JAC04231.1 CH479181 EDW31479.1 CCAG010011789 GFDF01010251 JAV03833.1 PYGN01001100 PSN37599.1 GFDL01006527 JAV28518.1 DS232215 EDS37740.1 CH477410 EAT41478.1 EAT41477.1 NEVH01017540 PNF24063.1 GEHC01001342 JAV46303.1 KK852779 KDR16706.1 CVRI01000059 CRL03226.1 APCN01000218 AAAB01008807 EAA04133.4 CRL03229.1 JXUM01037952 KQ561147 KXJ79566.1 AXCM01000153 AXCN02000591 CRL03225.1 CRL03224.1 UFQS01000574 UFQS01001061 UFQT01000574 UFQT01001061 SSX05038.1 SSX08771.1 SSX25400.1 GGFJ01010360 MBW59501.1 ATLV01012369 KE524785 KFB36642.1 EDO64517.1 GEZM01087520 JAV58651.1 KFB36643.1 CRL03227.1 GGFL01016173 MBW80351.1 LJIG01002325 KRT84615.1 GGFK01011121 MBW44442.1 ADMH02000160 ETN67531.1 KQ971354 EFA06001.1 GEBQ01010861 GEBQ01010679 JAT29116.1 JAT29298.1 GECZ01024131 JAS45638.1 GECU01036916 GECU01012548 JAS70790.1 JAS95158.1 GGFK01011232 MBW44553.1 APGK01025695 APGK01025696 KB740600 KB631982 ENN80111.1 ERL87597.1
RVE45017.1 AK401498 BAM18120.1 KQ459583 KPI98590.1 KQ459764 KPJ20104.1 JTDY01004797 KOB67742.1 AGBW02012100 OWR45673.1 OWR45672.1 CP012524 ALC41227.1 CH916367 EDW01594.1 CH902619 EDV35905.1 GAKP01014616 JAC44336.1 CH940648 EDW60085.1 CM000157 EDW89159.1 GBXI01002417 JAD11875.1 CM002911 KMY91936.1 AE013599 BT088861 AAF59278.1 ACS78061.1 CH954177 EDV59710.1 AY113626 AAM29631.1 CH933808 EDW08578.1 CH480816 EDW46733.1 CH963849 EDW74178.1 JXJN01030937 GDHF01021914 GDHF01015387 JAI30400.1 JAI36927.1 KA645096 AFP59725.1 JRES01000678 KNC29282.1 AJVK01010005 OUUW01000001 SPP73113.1 AJWK01011587 CM000071 EAL24775.1 GAMC01002325 JAC04231.1 CH479181 EDW31479.1 CCAG010011789 GFDF01010251 JAV03833.1 PYGN01001100 PSN37599.1 GFDL01006527 JAV28518.1 DS232215 EDS37740.1 CH477410 EAT41478.1 EAT41477.1 NEVH01017540 PNF24063.1 GEHC01001342 JAV46303.1 KK852779 KDR16706.1 CVRI01000059 CRL03226.1 APCN01000218 AAAB01008807 EAA04133.4 CRL03229.1 JXUM01037952 KQ561147 KXJ79566.1 AXCM01000153 AXCN02000591 CRL03225.1 CRL03224.1 UFQS01000574 UFQS01001061 UFQT01000574 UFQT01001061 SSX05038.1 SSX08771.1 SSX25400.1 GGFJ01010360 MBW59501.1 ATLV01012369 KE524785 KFB36642.1 EDO64517.1 GEZM01087520 JAV58651.1 KFB36643.1 CRL03227.1 GGFL01016173 MBW80351.1 LJIG01002325 KRT84615.1 GGFK01011121 MBW44442.1 ADMH02000160 ETN67531.1 KQ971354 EFA06001.1 GEBQ01010861 GEBQ01010679 JAT29116.1 JAT29298.1 GECZ01024131 JAS45638.1 GECU01036916 GECU01012548 JAS70790.1 JAS95158.1 GGFK01011232 MBW44553.1 APGK01025695 APGK01025696 KB740600 KB631982 ENN80111.1 ERL87597.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000037510
UP000007151
+ More
UP000092553 UP000001070 UP000007801 UP000008792 UP000095300 UP000002282 UP000000803 UP000008711 UP000009192 UP000001292 UP000007798 UP000192221 UP000092460 UP000095301 UP000037069 UP000092462 UP000268350 UP000092461 UP000091820 UP000092443 UP000001819 UP000092445 UP000078200 UP000008744 UP000092444 UP000245037 UP000002320 UP000008820 UP000235965 UP000027135 UP000183832 UP000075902 UP000075903 UP000075840 UP000007062 UP000069940 UP000249989 UP000075885 UP000075882 UP000075883 UP000075886 UP000076408 UP000075900 UP000030765 UP000076407 UP000075881 UP000075920 UP000075880 UP000075884 UP000000673 UP000007266 UP000069272 UP000192223 UP000019118 UP000030742
UP000092553 UP000001070 UP000007801 UP000008792 UP000095300 UP000002282 UP000000803 UP000008711 UP000009192 UP000001292 UP000007798 UP000192221 UP000092460 UP000095301 UP000037069 UP000092462 UP000268350 UP000092461 UP000091820 UP000092443 UP000001819 UP000092445 UP000078200 UP000008744 UP000092444 UP000245037 UP000002320 UP000008820 UP000235965 UP000027135 UP000183832 UP000075902 UP000075903 UP000075840 UP000007062 UP000069940 UP000249989 UP000075885 UP000075882 UP000075883 UP000075886 UP000076408 UP000075900 UP000030765 UP000076407 UP000075881 UP000075920 UP000075880 UP000075884 UP000000673 UP000007266 UP000069272 UP000192223 UP000019118 UP000030742
Interpro
Gene 3D
ProteinModelPortal
H9JWM4
A0A2W1BFM2
A0A2H1X291
A0A437B3E2
I4DJN0
A0A194Q5C0
+ More
A0A0N1IIM5 A0A0L7KWX8 A0A212EW14 A0A212EW05 A0A0M4EC44 B4J411 B3MI60 A0A034VLZ1 B4LN81 A0A1I8P7R3 B4P1M3 A0A0A1XLZ6 A0A0J9R7A4 A1Z6W5 B3NAF6 Q8MYS8 B4KU01 B4HQS3 B4MPY9 A0A1W4VDR6 A0A1B0ALD5 A0A1I8NDQ8 A0A0K8VDL3 T1P8K8 A0A0L0CAK1 A0A1B0D0N2 A0A3B0J2A9 A0A1B0CGU0 A0A1A9W135 A0A1A9X9J2 Q292T6 W8BS98 A0A1B0AAS6 A0A1A9UDF0 B4GCN1 A0A1B0FPB9 A0A1L8DBJ1 A0A2P8Y037 A0A1Q3FLX6 B0WZR5 Q174I0 Q174I1 A0A2J7Q653 A0A1W7R5A5 A0A067RAM3 A0A1J1IU46 A0A182U9D5 A0A182V2D3 A0A2C9GRU2 Q7QJT5 A0A1J1ISN1 A0A182G162 A0A182P9V9 A0A182KRW6 A0A182LRN2 A0A182Q5S8 A0A1J1IXE1 A0A1J1ISX4 A0A336M5U4 A0A182XZH4 A0A182MD23 A0A182XZH3 A0A182RFQ5 A0A2M4C2W3 A0A084VF98 A0A2C9GRW5 A7UR75 A0A182VIG8 A0A182TQP5 A0A182X6G4 A0A1Y1KE78 A0A182Q0A0 A0A182KDS1 A0A182WIT8 A0A084VF99 A0A182KRW4 A0A1J1ISL1 A0A182JAT8 A0A2M4DS39 A0A0T6BCH8 A0A2M4AUQ1 A0A182NHP0 W5JTS6 A0A182IS99 D6WR71 A0A1B6M049 A0A182FIB2 A0A1B6F5Z9 A0A1B6H815 A0A2M4AUU6 A0A1W4X938 N6UHN0
A0A0N1IIM5 A0A0L7KWX8 A0A212EW14 A0A212EW05 A0A0M4EC44 B4J411 B3MI60 A0A034VLZ1 B4LN81 A0A1I8P7R3 B4P1M3 A0A0A1XLZ6 A0A0J9R7A4 A1Z6W5 B3NAF6 Q8MYS8 B4KU01 B4HQS3 B4MPY9 A0A1W4VDR6 A0A1B0ALD5 A0A1I8NDQ8 A0A0K8VDL3 T1P8K8 A0A0L0CAK1 A0A1B0D0N2 A0A3B0J2A9 A0A1B0CGU0 A0A1A9W135 A0A1A9X9J2 Q292T6 W8BS98 A0A1B0AAS6 A0A1A9UDF0 B4GCN1 A0A1B0FPB9 A0A1L8DBJ1 A0A2P8Y037 A0A1Q3FLX6 B0WZR5 Q174I0 Q174I1 A0A2J7Q653 A0A1W7R5A5 A0A067RAM3 A0A1J1IU46 A0A182U9D5 A0A182V2D3 A0A2C9GRU2 Q7QJT5 A0A1J1ISN1 A0A182G162 A0A182P9V9 A0A182KRW6 A0A182LRN2 A0A182Q5S8 A0A1J1IXE1 A0A1J1ISX4 A0A336M5U4 A0A182XZH4 A0A182MD23 A0A182XZH3 A0A182RFQ5 A0A2M4C2W3 A0A084VF98 A0A2C9GRW5 A7UR75 A0A182VIG8 A0A182TQP5 A0A182X6G4 A0A1Y1KE78 A0A182Q0A0 A0A182KDS1 A0A182WIT8 A0A084VF99 A0A182KRW4 A0A1J1ISL1 A0A182JAT8 A0A2M4DS39 A0A0T6BCH8 A0A2M4AUQ1 A0A182NHP0 W5JTS6 A0A182IS99 D6WR71 A0A1B6M049 A0A182FIB2 A0A1B6F5Z9 A0A1B6H815 A0A2M4AUU6 A0A1W4X938 N6UHN0
PDB
2KG4
E-value=1.69238e-10,
Score=153
Ontologies
GO
PANTHER
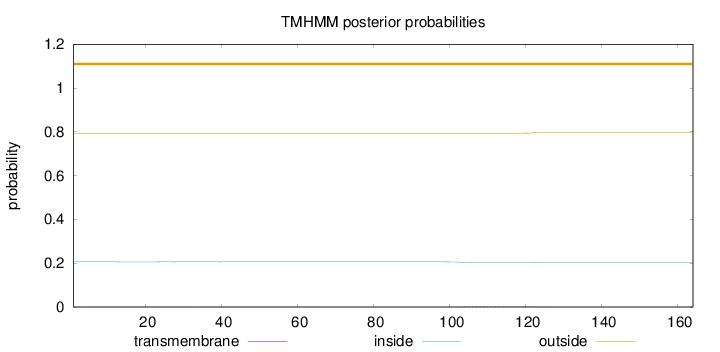
Topology
Length:
164
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08579
Exp number, first 60 AAs:
0.02082
Total prob of N-in:
0.20618
outside
1 - 164
Population Genetic Test Statistics
Pi
211.558878
Theta
215.697041
Tajima's D
-0.403125
CLR
0.629677
CSRT
0.263386830658467
Interpretation
Uncertain
