Gene
KWMTBOMO16168
Pre Gene Modal
BGIBMGA013951
Annotation
PREDICTED:_glucose_dehydrogenase_[FAD?_quinone]-like_[Papilio_polytes]
Location in the cell
Mitochondrial Reliability : 0.793
Sequence
CDS
ATGGTTTGGCAGCCAATAAATCTGGACCAAGCATGCCCGGGATCGACAAATCTAACTGCGTGCAGTTCCTTTGGGTACGTGTACTTGGACCTGCTGGTTCGACTCTTCGGTCAAACCAGCGGGAAGAGGAGACCTCCAGAGACCGAAGAGGAGAGAGAGTTTGACTTCATCGTTGTTGGAGCCGGTTCCGCTGGATGTGTTGTCGCCAATCGACTCAGTGCTGTAGGAAACTGGAAAGTACTACTCCTGGAAGCAGGAGACGAACAACCATTGGTGACCACAGTCCCGAGTCTGGCTATATCGCTGCTCCAGTCCAGCGTAGACTGGGCTTACCAGACTGAGCCGAATGGGAAGAGCTGCCTCGCCAGGCCGGGACAGCGCTGTAGCTGGCCGAGAGGCAAAACCATGGGCGGGTCTAGCTCTATAAACTCCTTAGCCTATGTTCGAGGGAACGAGCTGGACTACGACACTTGGGCTGCGAACGGAAACACAGGCTGGAGCTATAAAGAGGTCCTGCCTTACTTCATCAAGTCCGAGAAGAACTTGAACATCGCAGGTCTGAACCGGGAGTACCACGGCGTCGGCGGAGAGCAGTACGTCTCCAGATACCCGTACGTGGACCGACCGTCCATCATGATGACCGAGGCCTTCAACGAGAGGGGCGTCCCCATCGGAGACTTCAACGGCGAGAACCAGCTCAGCACGATGCAGTCGCAGACGTTCTCCTGCGGCGGGCAGCGCGTGTCCACCAACCGCGCCTTCATAGAGCCCGTCCGGGACAGGCGCCCCAACCTGCTCGTCAGGACCAACGCTGAAGTCCACAAGATACTGATCAATGAGAATAAGCAAGCGTACGGCGTGGTCTACAAGAGGAACTGCAAATGGCACACGGCCTACGCCAAGAAGGAGGTGATCATCAGCGCCGGCGCCATCAACTCGCCCAAACTACTTATGCTCTCGGGCATAGGACCTAAATACCACTTAGACGAGTTGGGCATAGAAGTGGTCAAAGATTTAGCCGTGGGTGAGAACCTGCACGACCACGTCACCTTCAACGGCATAATCGTGGCCCTGCCGAACGCGACGTCCACTAAAGTGAGCAACGACCAGCTGCTGAAAGAACTGTTTGCTTATAAATTGTCCCGACGCAAGAGAGGTCCGCTGGCGGCTAACGGGCCGACGAACTCCATCGCGTTTATAAGGACGGAGGCCGCGCTGCCGGCGCCCGACGTGCAGTTCCAAGTGGACAGCGTCACTTGGCAGGAGTACATCCGGCAGCCGACCCTCTACGAGAGCCTCACGATATTCCCATCCGCCTTCTACAACGCCCTGCTGCCGAGAACTATGAACTTGGTCCCGAAAAGTCGAGGCAAGTTGCTACTGAACCCAAAGAATCCTCGCGGAGCCCCACTACTCTACGCAAACTATTTCGACGACCCCACAGACTTGATTCCCCTAGTTAAGGGAGTCAAGTTCCTCCTGACGTTGGAGAACACGAAGGCGTTCAAACGTCTCGGCGCGTACTTCGTTAAGAAACCGTTGAAAGCTTGCAAGAATTATATTTGGGGCACTGACGACTACATTGTGTATTTGGCCAGATCTTATACTACGAGCACGTACCACCCGGTGGGAACGTGTAAAATGGGCCCGAAGTGGGATAAAAAAGCTGTTGTGGACCCACAATTGAGAGTCTATGGTGTTTCTGGTTTACGTGTCATTGATTCTTCTATAATGCCGTGTGCGCCGCGCGGGAACACCAACGCGCCATCTATTATGATCGGGGAGAGAGGAGTCGCTTTTGTTCTAAACTACTGGTTGAAGAAGTACAGCTAG
Protein
MVWQPINLDQACPGSTNLTACSSFGYVYLDLLVRLFGQTSGKRRPPETEEEREFDFIVVGAGSAGCVVANRLSAVGNWKVLLLEAGDEQPLVTTVPSLAISLLQSSVDWAYQTEPNGKSCLARPGQRCSWPRGKTMGGSSSINSLAYVRGNELDYDTWAANGNTGWSYKEVLPYFIKSEKNLNIAGLNREYHGVGGEQYVSRYPYVDRPSIMMTEAFNERGVPIGDFNGENQLSTMQSQTFSCGGQRVSTNRAFIEPVRDRRPNLLVRTNAEVHKILINENKQAYGVVYKRNCKWHTAYAKKEVIISAGAINSPKLLMLSGIGPKYHLDELGIEVVKDLAVGENLHDHVTFNGIIVALPNATSTKVSNDQLLKELFAYKLSRRKRGPLAANGPTNSIAFIRTEAALPAPDVQFQVDSVTWQEYIRQPTLYESLTIFPSAFYNALLPRTMNLVPKSRGKLLLNPKNPRGAPLLYANYFDDPTDLIPLVKGVKFLLTLENTKAFKRLGAYFVKKPLKACKNYIWGTDDYIVYLARSYTTSTYHPVGTCKMGPKWDKKAVVDPQLRVYGVSGLRVIDSSIMPCAPRGNTNAPSIMIGERGVAFVLNYWLKKYS
Summary
Cofactor
FAD
Similarity
Belongs to the GMC oxidoreductase family.
Uniprot
H9JWN7
A0A2H1X0I6
A0A0N1ICR5
A0A194Q0R9
A0A2W1BFE7
A0A437B3F2
+ More
A0A212EW05 A0A0L7KXH7 A0A437BP84 A0A194PY98 A0A0N1PFX8 H9JW74 A0A139WEV9 A0A212FGQ6 A0A212EVI0 U4V001 N6UNU0 U4TVS1 A0A212FGS2 A0A212EWQ4 A0A212FGU9 E0VFX6 A0A2A4JMY6 A0A2J7RBH7 A0A0L7LJQ0 A0A2P8ZKR6 A0A1W4WAJ1 A0A2H1WUZ8 H9JW75 A0A2S2QWK2 J9JJZ7 A0A2S2PRI8 A0A2W1BDQ5 A0A0M8ZT63 A0A0N0BBJ4 A0A154PBV5 A0A2A4JPK0 A0A2H1WV10 A0A3L8D6T0 A0A310ST84 A0A0M8ZRW9 E2BJK2 K7J043 A0A1W4WLT9 A0A232FLV0 A0A232EYA8 A0A088AJU8 A0A0L7RJT5 K7IPD7 A0A2A3EHZ9 A0A088AJU4 F4WAD9 A0A1A9XSX0 A0A1A9Z4T6 A0A1A9VSG2 B0WJG4 A0A3L8D5U9 A0A232EL32 A0A1A9WC57 A0A0C9RGK1 A0A0P6IJK2 A0A151I237 A0A1B0FJ02 K7J274 E0VUA7 D2A3N1 A0A151WMQ2 A0A232ETY1 A0A232F996 E2BJJ7 A0A0P5C063 A0A0P5M146 A0A0P5H6N8 A0A0P4YIW2 K7J278 A0A0P5SV55 A0A182H3F3 Q17DW0 A0A232F8Y5 A0A195E3M0 A0A0P6A2H0 A0A0P5UVJ4 A0A0P5C025 K7J0C5 A0A1I8M8L7 A0A0P4ZXT5 A0A0P5C863 A0A0P5I1Z9 A0A0P6JPQ5 A0A0P5H5Z6 A0A0P5JJB6 A0A2J7PRC3 A0A0P6GI86 A0A0P6GN41 A0A0P6FQ69 A0A0P5MS15 A0A0T6BDD9 A0A0P6GSC6 A0A0P5S474 A0A0P6AXG0
A0A212EW05 A0A0L7KXH7 A0A437BP84 A0A194PY98 A0A0N1PFX8 H9JW74 A0A139WEV9 A0A212FGQ6 A0A212EVI0 U4V001 N6UNU0 U4TVS1 A0A212FGS2 A0A212EWQ4 A0A212FGU9 E0VFX6 A0A2A4JMY6 A0A2J7RBH7 A0A0L7LJQ0 A0A2P8ZKR6 A0A1W4WAJ1 A0A2H1WUZ8 H9JW75 A0A2S2QWK2 J9JJZ7 A0A2S2PRI8 A0A2W1BDQ5 A0A0M8ZT63 A0A0N0BBJ4 A0A154PBV5 A0A2A4JPK0 A0A2H1WV10 A0A3L8D6T0 A0A310ST84 A0A0M8ZRW9 E2BJK2 K7J043 A0A1W4WLT9 A0A232FLV0 A0A232EYA8 A0A088AJU8 A0A0L7RJT5 K7IPD7 A0A2A3EHZ9 A0A088AJU4 F4WAD9 A0A1A9XSX0 A0A1A9Z4T6 A0A1A9VSG2 B0WJG4 A0A3L8D5U9 A0A232EL32 A0A1A9WC57 A0A0C9RGK1 A0A0P6IJK2 A0A151I237 A0A1B0FJ02 K7J274 E0VUA7 D2A3N1 A0A151WMQ2 A0A232ETY1 A0A232F996 E2BJJ7 A0A0P5C063 A0A0P5M146 A0A0P5H6N8 A0A0P4YIW2 K7J278 A0A0P5SV55 A0A182H3F3 Q17DW0 A0A232F8Y5 A0A195E3M0 A0A0P6A2H0 A0A0P5UVJ4 A0A0P5C025 K7J0C5 A0A1I8M8L7 A0A0P4ZXT5 A0A0P5C863 A0A0P5I1Z9 A0A0P6JPQ5 A0A0P5H5Z6 A0A0P5JJB6 A0A2J7PRC3 A0A0P6GI86 A0A0P6GN41 A0A0P6FQ69 A0A0P5MS15 A0A0T6BDD9 A0A0P6GSC6 A0A0P5S474 A0A0P6AXG0
Pubmed
EMBL
BABH01018352
ODYU01012451
SOQ58762.1
KQ459764
KPJ20105.1
KQ459583
+ More
KPI98589.1 KZ150184 PZC72475.1 RSAL01000177 RVE45018.1 AGBW02012100 OWR45672.1 JTDY01004797 KOB67741.1 RSAL01000025 RVE52179.1 KQ459588 KPI97724.1 KQ461158 KPJ08048.1 BABH01025443 KQ971354 KYB26474.1 AGBW02008601 OWR52931.1 AGBW02012199 OWR45467.1 KI208071 KI208851 ERL95783.1 ERL95945.1 APGK01024446 APGK01024447 KB740546 ENN80402.1 KB631740 ERL85684.1 OWR52932.1 AGBW02011908 OWR45938.1 OWR52933.1 DS235129 EEB12282.1 NWSH01000973 PCG73341.1 NEVH01005904 PNF38171.1 JTDY01000873 KOB75594.1 PYGN01000027 PSN57091.1 ODYU01011268 SOQ56899.1 GGMS01012934 MBY82137.1 ABLF02037931 GGMR01019368 MBY31987.1 KZ150399 PZC71020.1 KQ435889 KOX69466.1 KQ435991 KOX67668.1 KQ434869 KZC09322.1 PCG73342.1 ODYU01011271 SOQ56905.1 QOIP01000012 RLU16177.1 KQ759784 OAD62860.1 KQ435923 KOX68393.1 GL448571 EFN84159.1 NNAY01000068 OXU31327.1 NNAY01001663 OXU23279.1 KQ414579 KOC71130.1 KZ288238 PBC31350.1 GL888048 EGI68822.1 DS231960 EDS29160.1 RLU15835.1 NNAY01003669 OXU19031.1 GBYB01007495 JAG77262.1 GDIQ01010015 JAN84722.1 KQ976551 KYM80868.1 CCAG010013550 AAZX01003729 DS235784 EEB16963.1 KQ971348 EFA05531.1 KQ982937 KYQ49100.1 NNAY01002210 OXU21794.1 NNAY01000704 OXU26887.1 EFN84154.1 GDIP01177544 JAJ45858.1 GDIQ01247900 GDIQ01188610 GDIQ01164980 JAK86745.1 GDIQ01238146 JAK13579.1 GDIP01228120 JAI95281.1 AAZX01006810 AAZX01008181 GDIP01134833 LRGB01001348 JAL68881.1 KZS12804.1 JXUM01024549 KQ560694 KXJ81237.1 CH477290 EAT44641.1 NNAY01000675 OXU27062.1 KQ979701 KYN19666.1 GDIP01035901 JAM67814.1 GDIP01108083 JAL95631.1 GDIP01177551 JAJ45851.1 GDIP01228121 GDIP01219927 GDIP01174575 GDIP01158438 LRGB01002451 JAJ03475.1 KZS07507.1 GDIP01174531 JAJ48871.1 GDIQ01219255 JAK32470.1 GDIQ01001535 JAN93202.1 GDIQ01234793 GDIQ01145628 JAK16932.1 GDIQ01223788 JAK27937.1 NEVH01022362 PNF18877.1 GDIQ01032823 JAN61914.1 GDIQ01249546 GDIQ01198002 GDIQ01168260 GDIQ01164979 GDIQ01154367 GDIQ01146985 GDIQ01145627 GDIQ01080149 GDIQ01065407 GDIQ01032120 JAN62617.1 GDIQ01044718 JAN50019.1 GDIQ01174855 JAK76870.1 LJIG01001624 KRT85316.1 GDIQ01028962 JAN65775.1 GDIQ01092645 GDIQ01081757 JAL59081.1 GDIP01024006 JAM79709.1
KPI98589.1 KZ150184 PZC72475.1 RSAL01000177 RVE45018.1 AGBW02012100 OWR45672.1 JTDY01004797 KOB67741.1 RSAL01000025 RVE52179.1 KQ459588 KPI97724.1 KQ461158 KPJ08048.1 BABH01025443 KQ971354 KYB26474.1 AGBW02008601 OWR52931.1 AGBW02012199 OWR45467.1 KI208071 KI208851 ERL95783.1 ERL95945.1 APGK01024446 APGK01024447 KB740546 ENN80402.1 KB631740 ERL85684.1 OWR52932.1 AGBW02011908 OWR45938.1 OWR52933.1 DS235129 EEB12282.1 NWSH01000973 PCG73341.1 NEVH01005904 PNF38171.1 JTDY01000873 KOB75594.1 PYGN01000027 PSN57091.1 ODYU01011268 SOQ56899.1 GGMS01012934 MBY82137.1 ABLF02037931 GGMR01019368 MBY31987.1 KZ150399 PZC71020.1 KQ435889 KOX69466.1 KQ435991 KOX67668.1 KQ434869 KZC09322.1 PCG73342.1 ODYU01011271 SOQ56905.1 QOIP01000012 RLU16177.1 KQ759784 OAD62860.1 KQ435923 KOX68393.1 GL448571 EFN84159.1 NNAY01000068 OXU31327.1 NNAY01001663 OXU23279.1 KQ414579 KOC71130.1 KZ288238 PBC31350.1 GL888048 EGI68822.1 DS231960 EDS29160.1 RLU15835.1 NNAY01003669 OXU19031.1 GBYB01007495 JAG77262.1 GDIQ01010015 JAN84722.1 KQ976551 KYM80868.1 CCAG010013550 AAZX01003729 DS235784 EEB16963.1 KQ971348 EFA05531.1 KQ982937 KYQ49100.1 NNAY01002210 OXU21794.1 NNAY01000704 OXU26887.1 EFN84154.1 GDIP01177544 JAJ45858.1 GDIQ01247900 GDIQ01188610 GDIQ01164980 JAK86745.1 GDIQ01238146 JAK13579.1 GDIP01228120 JAI95281.1 AAZX01006810 AAZX01008181 GDIP01134833 LRGB01001348 JAL68881.1 KZS12804.1 JXUM01024549 KQ560694 KXJ81237.1 CH477290 EAT44641.1 NNAY01000675 OXU27062.1 KQ979701 KYN19666.1 GDIP01035901 JAM67814.1 GDIP01108083 JAL95631.1 GDIP01177551 JAJ45851.1 GDIP01228121 GDIP01219927 GDIP01174575 GDIP01158438 LRGB01002451 JAJ03475.1 KZS07507.1 GDIP01174531 JAJ48871.1 GDIQ01219255 JAK32470.1 GDIQ01001535 JAN93202.1 GDIQ01234793 GDIQ01145628 JAK16932.1 GDIQ01223788 JAK27937.1 NEVH01022362 PNF18877.1 GDIQ01032823 JAN61914.1 GDIQ01249546 GDIQ01198002 GDIQ01168260 GDIQ01164979 GDIQ01154367 GDIQ01146985 GDIQ01145627 GDIQ01080149 GDIQ01065407 GDIQ01032120 JAN62617.1 GDIQ01044718 JAN50019.1 GDIQ01174855 JAK76870.1 LJIG01001624 KRT85316.1 GDIQ01028962 JAN65775.1 GDIQ01092645 GDIQ01081757 JAL59081.1 GDIP01024006 JAM79709.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000007151
UP000037510
+ More
UP000007266 UP000030742 UP000019118 UP000009046 UP000218220 UP000235965 UP000245037 UP000192223 UP000007819 UP000053105 UP000076502 UP000279307 UP000008237 UP000002358 UP000215335 UP000005203 UP000053825 UP000242457 UP000007755 UP000092443 UP000092445 UP000078200 UP000002320 UP000091820 UP000078540 UP000092444 UP000075809 UP000076858 UP000069940 UP000249989 UP000008820 UP000078492 UP000095301
UP000007266 UP000030742 UP000019118 UP000009046 UP000218220 UP000235965 UP000245037 UP000192223 UP000007819 UP000053105 UP000076502 UP000279307 UP000008237 UP000002358 UP000215335 UP000005203 UP000053825 UP000242457 UP000007755 UP000092443 UP000092445 UP000078200 UP000002320 UP000091820 UP000078540 UP000092444 UP000075809 UP000076858 UP000069940 UP000249989 UP000008820 UP000078492 UP000095301
Interpro
SUPFAM
SSF51905
SSF51905
Gene 3D
CDD
ProteinModelPortal
H9JWN7
A0A2H1X0I6
A0A0N1ICR5
A0A194Q0R9
A0A2W1BFE7
A0A437B3F2
+ More
A0A212EW05 A0A0L7KXH7 A0A437BP84 A0A194PY98 A0A0N1PFX8 H9JW74 A0A139WEV9 A0A212FGQ6 A0A212EVI0 U4V001 N6UNU0 U4TVS1 A0A212FGS2 A0A212EWQ4 A0A212FGU9 E0VFX6 A0A2A4JMY6 A0A2J7RBH7 A0A0L7LJQ0 A0A2P8ZKR6 A0A1W4WAJ1 A0A2H1WUZ8 H9JW75 A0A2S2QWK2 J9JJZ7 A0A2S2PRI8 A0A2W1BDQ5 A0A0M8ZT63 A0A0N0BBJ4 A0A154PBV5 A0A2A4JPK0 A0A2H1WV10 A0A3L8D6T0 A0A310ST84 A0A0M8ZRW9 E2BJK2 K7J043 A0A1W4WLT9 A0A232FLV0 A0A232EYA8 A0A088AJU8 A0A0L7RJT5 K7IPD7 A0A2A3EHZ9 A0A088AJU4 F4WAD9 A0A1A9XSX0 A0A1A9Z4T6 A0A1A9VSG2 B0WJG4 A0A3L8D5U9 A0A232EL32 A0A1A9WC57 A0A0C9RGK1 A0A0P6IJK2 A0A151I237 A0A1B0FJ02 K7J274 E0VUA7 D2A3N1 A0A151WMQ2 A0A232ETY1 A0A232F996 E2BJJ7 A0A0P5C063 A0A0P5M146 A0A0P5H6N8 A0A0P4YIW2 K7J278 A0A0P5SV55 A0A182H3F3 Q17DW0 A0A232F8Y5 A0A195E3M0 A0A0P6A2H0 A0A0P5UVJ4 A0A0P5C025 K7J0C5 A0A1I8M8L7 A0A0P4ZXT5 A0A0P5C863 A0A0P5I1Z9 A0A0P6JPQ5 A0A0P5H5Z6 A0A0P5JJB6 A0A2J7PRC3 A0A0P6GI86 A0A0P6GN41 A0A0P6FQ69 A0A0P5MS15 A0A0T6BDD9 A0A0P6GSC6 A0A0P5S474 A0A0P6AXG0
A0A212EW05 A0A0L7KXH7 A0A437BP84 A0A194PY98 A0A0N1PFX8 H9JW74 A0A139WEV9 A0A212FGQ6 A0A212EVI0 U4V001 N6UNU0 U4TVS1 A0A212FGS2 A0A212EWQ4 A0A212FGU9 E0VFX6 A0A2A4JMY6 A0A2J7RBH7 A0A0L7LJQ0 A0A2P8ZKR6 A0A1W4WAJ1 A0A2H1WUZ8 H9JW75 A0A2S2QWK2 J9JJZ7 A0A2S2PRI8 A0A2W1BDQ5 A0A0M8ZT63 A0A0N0BBJ4 A0A154PBV5 A0A2A4JPK0 A0A2H1WV10 A0A3L8D6T0 A0A310ST84 A0A0M8ZRW9 E2BJK2 K7J043 A0A1W4WLT9 A0A232FLV0 A0A232EYA8 A0A088AJU8 A0A0L7RJT5 K7IPD7 A0A2A3EHZ9 A0A088AJU4 F4WAD9 A0A1A9XSX0 A0A1A9Z4T6 A0A1A9VSG2 B0WJG4 A0A3L8D5U9 A0A232EL32 A0A1A9WC57 A0A0C9RGK1 A0A0P6IJK2 A0A151I237 A0A1B0FJ02 K7J274 E0VUA7 D2A3N1 A0A151WMQ2 A0A232ETY1 A0A232F996 E2BJJ7 A0A0P5C063 A0A0P5M146 A0A0P5H6N8 A0A0P4YIW2 K7J278 A0A0P5SV55 A0A182H3F3 Q17DW0 A0A232F8Y5 A0A195E3M0 A0A0P6A2H0 A0A0P5UVJ4 A0A0P5C025 K7J0C5 A0A1I8M8L7 A0A0P4ZXT5 A0A0P5C863 A0A0P5I1Z9 A0A0P6JPQ5 A0A0P5H5Z6 A0A0P5JJB6 A0A2J7PRC3 A0A0P6GI86 A0A0P6GN41 A0A0P6FQ69 A0A0P5MS15 A0A0T6BDD9 A0A0P6GSC6 A0A0P5S474 A0A0P6AXG0
PDB
5NCC
E-value=5.68766e-53,
Score=527
Ontologies
GO
Topology
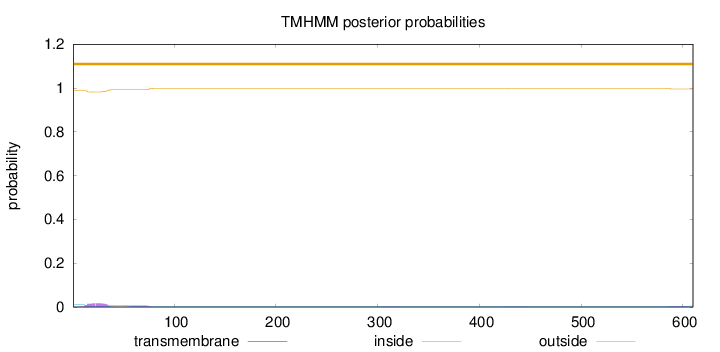
Length:
610
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.545660000000001
Exp number, first 60 AAs:
0.38036
Total prob of N-in:
0.01223
outside
1 - 610
Population Genetic Test Statistics
Pi
180.375208
Theta
27.014284
Tajima's D
1.273653
CLR
0.025568
CSRT
0.739713014349282
Interpretation
Uncertain
