Gene
KWMTBOMO16165
Pre Gene Modal
BGIBMGA013948
Annotation
PREDICTED:_homeobox_protein_orthopedia_[Bombyx_mori]
Full name
Homeobox protein orthopedia
Location in the cell
Nuclear Reliability : 3.269
Sequence
CDS
ATGGTCTGGTTCCAGAACAGACGGGCGAAGTGGAAAAAGCGCAAGAAGAGCACCAATGTGTTTAGATCTCCAGGAGCGTTGCTTCCTTCACACGGCTTACCACCGTTCGGGGCTGGGGACGTCTGTGGACCTGGTGTCTTTGCAGACAGACCTTGGTCGATGCCCGCTGGTCTTGGGCAACTATCGCAGTCGGGAGTGTCTATGGGAGGTTTCGGTGCGGGCACCCTGCCGCAGCTCGCCGCGGACCTGAACCCCTCGCTCCCCATCAGCTCAGTGGCGTCGGGACAACCTTACCAGGGACACTATCCGCTGAACTCTTTAGGAGACACGATGATGTCCTACTGCAACAACATAGCGAGCGGACAACAGGCCTTGGAGGGCTCCCCCACCCCCCCGCACATGGGACACTGCGCCAACAACAACGCGTCGCCTGACGCTTCAGGTAAACTTGCCAGGATCAAGTCAATTCGCAACTTTTCACCGGTGGTAGGACCTTTTGTGAGTCCGCGCGGGTAG
Protein
MVWFQNRRAKWKKRKKSTNVFRSPGALLPSHGLPPFGAGDVCGPGVFADRPWSMPAGLGQLSQSGVSMGGFGAGTLPQLAADLNPSLPISSVASGQPYQGHYPLNSLGDTMMSYCNNIASGQQALEGSPTPPHMGHCANNNASPDASGKLARIKSIRNFSPVVGPFVSPRG
Summary
Description
May be involved in the development of the central nervous system.
Similarity
Belongs to the paired homeobox family.
Belongs to the paired homeobox family. Bicoid subfamily.
Belongs to the paired homeobox family. Bicoid subfamily.
Keywords
Alternative splicing
Complete proteome
Developmental protein
DNA-binding
Homeobox
Nucleus
Reference proteome
Feature
chain Homeobox protein orthopedia
splice variant In isoform C.
splice variant In isoform C.
Uniprot
H9JWN4
A0A2A4K001
A0A2H1WU31
A0A2W1BMD9
A0A437B3I6
A0A212FHC3
+ More
A0A0N0PEJ3 A0A194Q0R6 A0A1W4WKM7 A0A1W4WAK1 A0A1W4WLM0 A0A1W4WAH8 V5IA52 A0A2J7PF19 A0A0L7RBT3 A0A1D2N5S5 B3MIQ9 A0A0A1XRQ9 A0A1I8MBU0 B4NNJ8 A0A226EZT7 A0A182FCF0 B4J985 B4KQM5 A0A1I8PSX6 A0A1L8DDA5 W5JUX1 B4I7G0 A0A0J9U721 A0A0B4LFZ2 A0A0J9RHI9 A0A0B4LFU7 A0A0B4LH19 B4QFI5 P56672 A0A0J9RIP0 B3NK04 A0A3B0JER8 B4PA05 Q28ZR5 A0A1W4VHJ1 B4GGW2 A0A3B0IZY6 A0A182N895 A0A3B0JK11 A0A182RTL8 A0A182LBA4 A0A182HP95 A0A182WIJ4 A0A182MI15 A0A1S4EZJ8 A0A182X492 A0A182P6B1 A0A182VGE8 A0A0L0CFV8 A0A1A9VY58 A0A182JDG6 Q17KG1 A0A1A9XJ95 A0A1A9ZNN7 A0A1B0B220 A0A1B0FDS8 A0A1A9WMS6 A0A1J1IUX4 A0A154NX82 A0A336MRB6 A0A336MG30 A0A0C9R705 A0A182TDZ5 A0A0A1X6P7 A0A0A1XHT5
A0A0N0PEJ3 A0A194Q0R6 A0A1W4WKM7 A0A1W4WAK1 A0A1W4WLM0 A0A1W4WAH8 V5IA52 A0A2J7PF19 A0A0L7RBT3 A0A1D2N5S5 B3MIQ9 A0A0A1XRQ9 A0A1I8MBU0 B4NNJ8 A0A226EZT7 A0A182FCF0 B4J985 B4KQM5 A0A1I8PSX6 A0A1L8DDA5 W5JUX1 B4I7G0 A0A0J9U721 A0A0B4LFZ2 A0A0J9RHI9 A0A0B4LFU7 A0A0B4LH19 B4QFI5 P56672 A0A0J9RIP0 B3NK04 A0A3B0JER8 B4PA05 Q28ZR5 A0A1W4VHJ1 B4GGW2 A0A3B0IZY6 A0A182N895 A0A3B0JK11 A0A182RTL8 A0A182LBA4 A0A182HP95 A0A182WIJ4 A0A182MI15 A0A1S4EZJ8 A0A182X492 A0A182P6B1 A0A182VGE8 A0A0L0CFV8 A0A1A9VY58 A0A182JDG6 Q17KG1 A0A1A9XJ95 A0A1A9ZNN7 A0A1B0B220 A0A1B0FDS8 A0A1A9WMS6 A0A1J1IUX4 A0A154NX82 A0A336MRB6 A0A336MG30 A0A0C9R705 A0A182TDZ5 A0A0A1X6P7 A0A0A1XHT5
Pubmed
EMBL
BABH01018342
BABH01018343
NWSH01000308
PCG77567.1
ODYU01011064
SOQ56563.1
+ More
KZ149970 PZC76098.1 RSAL01000177 RVE45020.1 AGBW02008507 OWR53135.1 KQ459764 KPJ20109.1 KQ459583 KPI98584.1 GALX01001790 JAB66676.1 NEVH01026086 PNF14927.1 KQ414617 KOC68397.1 LJIJ01000197 ODN00604.1 CH902619 EDV35969.2 GBXI01000630 JAD13662.1 CH964282 EDW85937.2 LNIX01000001 OXA62401.1 CH916367 EDW02460.1 CH933808 EDW08194.2 GFDF01009652 JAV04432.1 ADMH02000391 ETN66745.1 CH480824 EDW56535.1 CM002911 KMY95360.1 AE013599 AHN56445.1 KMY95357.1 AHN56446.1 AHN56449.1 CM000362 EDX07977.1 KMY95353.1 BT023887 BT030422 KMY95359.1 CH954179 EDV55370.2 OUUW01000001 SPP73790.1 CM000158 EDW91336.2 CM000071 EAL25546.3 CH479183 EDW35732.1 SPP73787.1 SPP73789.1 APCN01002931 AXCM01004441 JRES01000440 KNC31130.1 CH477224 EAT47190.1 JXJN01007394 CCAG010007043 CVRI01000059 CRL03506.1 KQ434777 KZC04172.1 UFQT01002088 SSX32620.1 UFQS01000530 UFQT01000530 SSX04675.1 SSX25038.1 GBYB01008582 JAG78349.1 GBXI01007293 JAD06999.1 GBXI01004224 JAD10068.1
KZ149970 PZC76098.1 RSAL01000177 RVE45020.1 AGBW02008507 OWR53135.1 KQ459764 KPJ20109.1 KQ459583 KPI98584.1 GALX01001790 JAB66676.1 NEVH01026086 PNF14927.1 KQ414617 KOC68397.1 LJIJ01000197 ODN00604.1 CH902619 EDV35969.2 GBXI01000630 JAD13662.1 CH964282 EDW85937.2 LNIX01000001 OXA62401.1 CH916367 EDW02460.1 CH933808 EDW08194.2 GFDF01009652 JAV04432.1 ADMH02000391 ETN66745.1 CH480824 EDW56535.1 CM002911 KMY95360.1 AE013599 AHN56445.1 KMY95357.1 AHN56446.1 AHN56449.1 CM000362 EDX07977.1 KMY95353.1 BT023887 BT030422 KMY95359.1 CH954179 EDV55370.2 OUUW01000001 SPP73790.1 CM000158 EDW91336.2 CM000071 EAL25546.3 CH479183 EDW35732.1 SPP73787.1 SPP73789.1 APCN01002931 AXCM01004441 JRES01000440 KNC31130.1 CH477224 EAT47190.1 JXJN01007394 CCAG010007043 CVRI01000059 CRL03506.1 KQ434777 KZC04172.1 UFQT01002088 SSX32620.1 UFQS01000530 UFQT01000530 SSX04675.1 SSX25038.1 GBYB01008582 JAG78349.1 GBXI01007293 JAD06999.1 GBXI01004224 JAD10068.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
+ More
UP000192223 UP000235965 UP000053825 UP000094527 UP000007801 UP000095301 UP000007798 UP000198287 UP000069272 UP000001070 UP000009192 UP000095300 UP000000673 UP000001292 UP000000803 UP000000304 UP000008711 UP000268350 UP000002282 UP000001819 UP000192221 UP000008744 UP000075884 UP000075900 UP000075882 UP000075840 UP000075920 UP000075883 UP000076407 UP000075885 UP000075903 UP000037069 UP000078200 UP000075880 UP000008820 UP000092443 UP000092445 UP000092460 UP000092444 UP000091820 UP000183832 UP000076502 UP000075902
UP000192223 UP000235965 UP000053825 UP000094527 UP000007801 UP000095301 UP000007798 UP000198287 UP000069272 UP000001070 UP000009192 UP000095300 UP000000673 UP000001292 UP000000803 UP000000304 UP000008711 UP000268350 UP000002282 UP000001819 UP000192221 UP000008744 UP000075884 UP000075900 UP000075882 UP000075840 UP000075920 UP000075883 UP000076407 UP000075885 UP000075903 UP000037069 UP000078200 UP000075880 UP000008820 UP000092443 UP000092445 UP000092460 UP000092444 UP000091820 UP000183832 UP000076502 UP000075902
Pfam
PF00046 Homeodomain
Interpro
SUPFAM
SSF46689
SSF46689
ProteinModelPortal
H9JWN4
A0A2A4K001
A0A2H1WU31
A0A2W1BMD9
A0A437B3I6
A0A212FHC3
+ More
A0A0N0PEJ3 A0A194Q0R6 A0A1W4WKM7 A0A1W4WAK1 A0A1W4WLM0 A0A1W4WAH8 V5IA52 A0A2J7PF19 A0A0L7RBT3 A0A1D2N5S5 B3MIQ9 A0A0A1XRQ9 A0A1I8MBU0 B4NNJ8 A0A226EZT7 A0A182FCF0 B4J985 B4KQM5 A0A1I8PSX6 A0A1L8DDA5 W5JUX1 B4I7G0 A0A0J9U721 A0A0B4LFZ2 A0A0J9RHI9 A0A0B4LFU7 A0A0B4LH19 B4QFI5 P56672 A0A0J9RIP0 B3NK04 A0A3B0JER8 B4PA05 Q28ZR5 A0A1W4VHJ1 B4GGW2 A0A3B0IZY6 A0A182N895 A0A3B0JK11 A0A182RTL8 A0A182LBA4 A0A182HP95 A0A182WIJ4 A0A182MI15 A0A1S4EZJ8 A0A182X492 A0A182P6B1 A0A182VGE8 A0A0L0CFV8 A0A1A9VY58 A0A182JDG6 Q17KG1 A0A1A9XJ95 A0A1A9ZNN7 A0A1B0B220 A0A1B0FDS8 A0A1A9WMS6 A0A1J1IUX4 A0A154NX82 A0A336MRB6 A0A336MG30 A0A0C9R705 A0A182TDZ5 A0A0A1X6P7 A0A0A1XHT5
A0A0N0PEJ3 A0A194Q0R6 A0A1W4WKM7 A0A1W4WAK1 A0A1W4WLM0 A0A1W4WAH8 V5IA52 A0A2J7PF19 A0A0L7RBT3 A0A1D2N5S5 B3MIQ9 A0A0A1XRQ9 A0A1I8MBU0 B4NNJ8 A0A226EZT7 A0A182FCF0 B4J985 B4KQM5 A0A1I8PSX6 A0A1L8DDA5 W5JUX1 B4I7G0 A0A0J9U721 A0A0B4LFZ2 A0A0J9RHI9 A0A0B4LFU7 A0A0B4LH19 B4QFI5 P56672 A0A0J9RIP0 B3NK04 A0A3B0JER8 B4PA05 Q28ZR5 A0A1W4VHJ1 B4GGW2 A0A3B0IZY6 A0A182N895 A0A3B0JK11 A0A182RTL8 A0A182LBA4 A0A182HP95 A0A182WIJ4 A0A182MI15 A0A1S4EZJ8 A0A182X492 A0A182P6B1 A0A182VGE8 A0A0L0CFV8 A0A1A9VY58 A0A182JDG6 Q17KG1 A0A1A9XJ95 A0A1A9ZNN7 A0A1B0B220 A0A1B0FDS8 A0A1A9WMS6 A0A1J1IUX4 A0A154NX82 A0A336MRB6 A0A336MG30 A0A0C9R705 A0A182TDZ5 A0A0A1X6P7 A0A0A1XHT5
Ontologies
GO
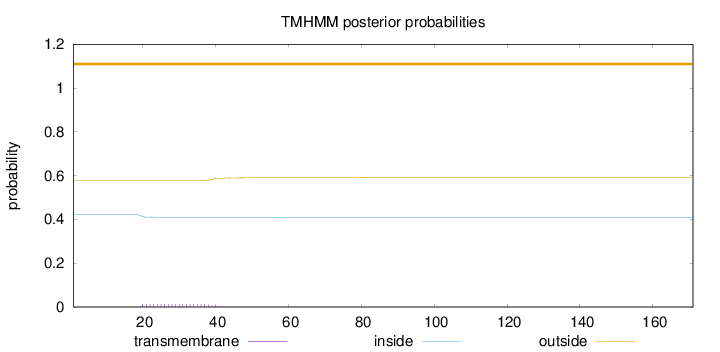
Topology
Subcellular location
Nucleus
Length:
171
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.277650000000001
Exp number, first 60 AAs:
0.25999
Total prob of N-in:
0.42209
outside
1 - 171
Population Genetic Test Statistics
Pi
195.03324
Theta
185.666555
Tajima's D
2.011714
CLR
0.352756
CSRT
0.879406029698515
Interpretation
Uncertain
