Gene
KWMTBOMO16159 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013945
Annotation
actin?_partial_[Zygaena_filipendulae]
Full name
Actin, muscle
+ More
Actin, muscle-type A1
Actin, muscle-type A1
Location in the cell
Cytoplasmic Reliability : 3.869
Sequence
CDS
ATGTGCGACGACGATGTTGCTGCGCTTGTAGTCGACAATGGCTCCGGCATGTGCAAGGCCGGTTTCGCCGGCGACGACGCGCCCCGCGCCGTCTTCCCGTCCATCGTGGGTCGCCCTCGCCACCAGGGCGTGATGGTCGGTATGGGCCAGAAAGACTCCTACGTGGGTGACGAGGCCCAGAGCAAGAGAGGTATCCTCACTCTGAAGTACCCCATCGAGCACGGTATCATCACCAACTGGGATGACATGGAGAAGATCTGGCACCACACCTTCTACAATGAGCTGCGTGTCGCCCCCGAGGAGCACCCCGTGCTGCTCACCGAAGCCCCCCTCAACCCCAAGGCCAACAGGGAGAAGATGACCCAGATCATGTTTGAGACCTTCAACTCCCCCGCCATGTACGTCGCCATCCAGGCCGTGCTCTCGCTGTACGCCTCCGGTCGTACCACCGGTATCGTGCTCGACTCCGGAGATGGTGTCTCCCACACCGTACCCATCTACGAAGGTTACGCTCTGCCCCACGCCATCCTCCGTCTGGACTTGGCTGGTCGCGACTTGACCGACTACCTCATGAAGATCCTCACCGAGAGGGGTTACTCGTTCACCACCACCGCTGAGAGGGAAATCGTTCGTGACATCAAGGAGAAGCTCTGCTATGTCGCCCTCGACTTCGAGCAGGAGATGGCCACCGCTGCCGCCTCCACCTCCCTCGAGAAGTCCTACGAACTTCCTGACGGTCAAGTGATCACCATCGGTAACGAGAGGTTCCGTTGTCCCGAGGCTCTCTTCCAGCCTTCCTTCCTGGGTATGGAATCGTGCGGCATCCACGAGACCGTGTACAACTCCATCATGAAGTGCGACGTCGACATCCGTAAGGACCTGTACGCCAACACCGTCATGTCCGGTGGTACCACCATGTACCCCGGTATCGCCGACAGGATGCAGAAGGAGATCACCGCCCTCGCGCCCTCGACCATCAAGATCAAGATCATCGCTCCCCCCGAGAGGAAGTACTCCGTATGGATCGGTGGATCCATCCTGGCTTCCCTGTCCACCTTCCAGCAGATGTGGATCTCGAAGGAGGAATACGACGAGTCCGGCCCCGGCATCGTCCACCGCAAGTGCTTCTAA
Protein
MCDDDVAALVVDNGSGMCKAGFAGDDAPRAVFPSIVGRPRHQGVMVGMGQKDSYVGDEAQSKRGILTLKYPIEHGIITNWDDMEKIWHHTFYNELRVAPEEHPVLLTEAPLNPKANREKMTQIMFETFNSPAMYVAIQAVLSLYASGRTTGIVLDSGDGVSHTVPIYEGYALPHAILRLDLAGRDLTDYLMKILTERGYSFTTTAEREIVRDIKEKLCYVALDFEQEMATAAASTSLEKSYELPDGQVITIGNERFRCPEALFQPSFLGMESCGIHETVYNSIMKCDVDIRKDLYANTVMSGGTTMYPGIADRMQKEITALAPSTIKIKIIAPPERKYSVWIGGSILASLSTFQQMWISKEEYDESGPGIVHRKCF
Summary
Subunit
Polymerization of globular actin (G-actin) leads to a structural filament (F-actin) in the form of a two-stranded helix. Each actin can bind to 4 others.
Miscellaneous
There are at least 5 different actin genes in this organism.
Similarity
Belongs to the actin family.
Keywords
Acetylation
ATP-binding
Cytoplasm
Cytoskeleton
Muscle protein
Nucleotide-binding
Oxidation
Complete proteome
Reference proteome
Feature
propeptide Removed in mature form
chain Actin, muscle
chain Actin, muscle
Uniprot
X5CT98
V9NEM2
A0A0L7LTL0
A0A2W1BR20
A0A2A4JIS6
I4DLT1
+ More
A0A0N1IAY6 A0A2Z5STE6 B2Z6M6 T1DFC0 U5EUS3 A0A1W6S692 A0A2H1X0F8 A0A182YFN1 H9JWN1 A0A291FJB0 A0A2Z4N445 A0A212EZ57 A0A182P6B7 A0A182M762 A0A182PZM2 A0A182V6G0 A0A182N891 A0A182U5H7 A0A182WQH5 B0XB41 Q17KG3 A0A084W2J0 A0A182LB97 A0A182ITR0 A0A182X497 A8B1J8 A0A182FZK9 A0JCK7 A0A182RTL3 A0A182HPA0 A0A023ES50 P49871 B2DBP0 B0LF73 A0A1B0D7H8 A0A151JMQ0 A0A151I4W0 K7J803 A0A195FX88 A0A158NXU2 A0A3G1SUZ3 A0A151WIN4 Q6ELZ7 Q45L89 D3KU63 A0A1W4XH28 A0A2Z4G633 A7WPD3 A0A0P5CGV3 A0A026X3P0 E2IV57 P07836 F4WLU8 A0A1W4XWB9 A0A2R4GMB1 F2D4P0 A0A224XRR1 Q5XXT4 Q3HNA6 Q5RLJ4 A0A125RA11 A0A2D1QW10 A0A2H8TVL9 V5GR51 A0A1S3CVP9 A0A3G4R8Q5 A0A3G1DKW0 A0A023J916 A0A0P4VVE9 A0A140EFS7 Q5XUA4 D6WF19 A0A0V0G386 X1X262 A0A170ZGA9 Q6PPI5 A0A2J7RHY5 A1BPP6 A0A067QL86 M9VDW4 A0A1S5RIQ2 A0A023VZM4 A0A2Z5RE76 A0A125RA09 A0A1Y1LUA6 R4FQA8 N6TH73 A0A1B1UZE0 A0A0P5CHV2 A0A1B6EXE0 E0VKP4 A0A1B6CK57 A0A0A9W354 A0A0K2UPC6
A0A0N1IAY6 A0A2Z5STE6 B2Z6M6 T1DFC0 U5EUS3 A0A1W6S692 A0A2H1X0F8 A0A182YFN1 H9JWN1 A0A291FJB0 A0A2Z4N445 A0A212EZ57 A0A182P6B7 A0A182M762 A0A182PZM2 A0A182V6G0 A0A182N891 A0A182U5H7 A0A182WQH5 B0XB41 Q17KG3 A0A084W2J0 A0A182LB97 A0A182ITR0 A0A182X497 A8B1J8 A0A182FZK9 A0JCK7 A0A182RTL3 A0A182HPA0 A0A023ES50 P49871 B2DBP0 B0LF73 A0A1B0D7H8 A0A151JMQ0 A0A151I4W0 K7J803 A0A195FX88 A0A158NXU2 A0A3G1SUZ3 A0A151WIN4 Q6ELZ7 Q45L89 D3KU63 A0A1W4XH28 A0A2Z4G633 A7WPD3 A0A0P5CGV3 A0A026X3P0 E2IV57 P07836 F4WLU8 A0A1W4XWB9 A0A2R4GMB1 F2D4P0 A0A224XRR1 Q5XXT4 Q3HNA6 Q5RLJ4 A0A125RA11 A0A2D1QW10 A0A2H8TVL9 V5GR51 A0A1S3CVP9 A0A3G4R8Q5 A0A3G1DKW0 A0A023J916 A0A0P4VVE9 A0A140EFS7 Q5XUA4 D6WF19 A0A0V0G386 X1X262 A0A170ZGA9 Q6PPI5 A0A2J7RHY5 A1BPP6 A0A067QL86 M9VDW4 A0A1S5RIQ2 A0A023VZM4 A0A2Z5RE76 A0A125RA09 A0A1Y1LUA6 R4FQA8 N6TH73 A0A1B1UZE0 A0A0P5CHV2 A0A1B6EXE0 E0VKP4 A0A1B6CK57 A0A0A9W354 A0A0K2UPC6
Pubmed
26227816
28756777
22651552
26354079
25244985
19121390
+ More
22118469 17510324 24438588 20966253 12364791 14747013 17210077 24945155 26483478 8344462 20075255 21347285 12429127 20022415 29889877 24508170 3562238 21719571 29590372 21415278 16537425 26826390 27483241 27129103 26873291 18362917 19820115 17469070 24845553 27494615 26760975 28004739 23537049 20566863 25401762
22118469 17510324 24438588 20966253 12364791 14747013 17210077 24945155 26483478 8344462 20075255 21347285 12429127 20022415 29889877 24508170 3562238 21719571 29590372 21415278 16537425 26826390 27483241 27129103 26873291 18362917 19820115 17469070 24845553 27494615 26760975 28004739 23537049 20566863 25401762
EMBL
KJ192330
AHW40461.1
KF022227
AGN98097.1
JTDY01000114
KOB78777.1
+ More
KZ149970 PZC76094.1 NWSH01001327 PCG71676.1 AK402249 BAM18871.1 KQ459764 KPJ20112.1 LC328974 BBA78421.1 EU601172 ACD40028.1 GAMD01003191 JAA98399.1 GANO01001336 JAB58535.1 KY231202 ARO75867.1 ODYU01012432 SOQ58732.1 BABH01018323 KY039972 ATG34151.1 MG132199 AWX63637.1 AGBW02011390 OWR46741.1 AXCM01013032 AXCN02000961 DS232612 EDS44094.1 CH477224 EAT47188.1 ATLV01019621 KE525275 KFB44434.1 AXCP01003441 AAAB01008900 EAA09436.4 JN410820 KU872540 AB282645 AEP25396.1 AOM81402.1 BAF36825.1 APCN01002933 JXUM01017643 GAPW01002037 KQ560489 JAC11561.1 KXJ82141.1 L13764 AB264725 BAG30799.1 EU035314 ABW03225.1 AJVK01026919 KQ978931 KYN27466.1 KQ976462 KYM84633.1 AAZX01005009 KQ981204 KYN44932.1 ADTU01003540 ADTU01003541 MG243593 AXU38338.1 KQ983089 KYQ47686.1 AY289764 AAQ24506.1 DQ124691 AAZ31061.1 AB509262 BAI77996.1 MG983770 AWV96605.1 AM850110 CAO98769.1 GDIP01170424 JAJ52978.1 KK107013 EZA62930.1 HM629437 HM629439 HM629441 ADN84927.1 X05185 GL888216 EGI64830.1 MG264369 AVT69429.1 AK358850 BAJ90061.1 GFTR01005593 JAW10833.1 AY725778 AAU95191.1 DQ206832 ABA62321.1 AY817141 AAV65298.1 KU196670 AMD16550.1 KY038869 ATP16752.1 GFXV01006509 MBW18314.1 GALX01004399 JAB64067.1 MG993326 AYU58903.1 KU188517 ANE06405.1 KF792064 AHH25009.1 GDKW01000574 JAI56021.1 KT369806 AML25535.1 AY737550 AAU84943.1 KQ971319 EFA00468.1 GECL01003587 JAP02537.1 ABLF02032889 GEMB01002235 JAS00945.1 AY588061 AAT01073.1 NEVH01003508 PNF40452.1 DQ450899 ABE27979.1 KK853207 KDR09838.1 KC161211 AGJ76539.1 KX108734 ANV22067.1 KJ507199 AHY22050.1 FX983744 BAX07243.1 KU196668 AMD16548.1 GEZM01048387 GEZM01048386 JAV76591.1 ACPB03021529 GAHY01000509 JAA77001.1 APGK01027120 KB740648 KB632292 ENN79784.1 ERL91608.1 KU521368 KX428475 ANS71248.1 ANW09586.1 GDIP01175047 GDIP01170376 JAJ53026.1 GECZ01027298 GECZ01011797 JAS42471.1 JAS57972.1 DS235250 EEB13950.1 GEDC01023461 GEDC01016965 GEDC01002583 JAS13837.1 JAS20333.1 JAS34715.1 GBHO01044314 GBRD01005390 JAF99289.1 JAG60431.1 HACA01022742 CDW40103.1
KZ149970 PZC76094.1 NWSH01001327 PCG71676.1 AK402249 BAM18871.1 KQ459764 KPJ20112.1 LC328974 BBA78421.1 EU601172 ACD40028.1 GAMD01003191 JAA98399.1 GANO01001336 JAB58535.1 KY231202 ARO75867.1 ODYU01012432 SOQ58732.1 BABH01018323 KY039972 ATG34151.1 MG132199 AWX63637.1 AGBW02011390 OWR46741.1 AXCM01013032 AXCN02000961 DS232612 EDS44094.1 CH477224 EAT47188.1 ATLV01019621 KE525275 KFB44434.1 AXCP01003441 AAAB01008900 EAA09436.4 JN410820 KU872540 AB282645 AEP25396.1 AOM81402.1 BAF36825.1 APCN01002933 JXUM01017643 GAPW01002037 KQ560489 JAC11561.1 KXJ82141.1 L13764 AB264725 BAG30799.1 EU035314 ABW03225.1 AJVK01026919 KQ978931 KYN27466.1 KQ976462 KYM84633.1 AAZX01005009 KQ981204 KYN44932.1 ADTU01003540 ADTU01003541 MG243593 AXU38338.1 KQ983089 KYQ47686.1 AY289764 AAQ24506.1 DQ124691 AAZ31061.1 AB509262 BAI77996.1 MG983770 AWV96605.1 AM850110 CAO98769.1 GDIP01170424 JAJ52978.1 KK107013 EZA62930.1 HM629437 HM629439 HM629441 ADN84927.1 X05185 GL888216 EGI64830.1 MG264369 AVT69429.1 AK358850 BAJ90061.1 GFTR01005593 JAW10833.1 AY725778 AAU95191.1 DQ206832 ABA62321.1 AY817141 AAV65298.1 KU196670 AMD16550.1 KY038869 ATP16752.1 GFXV01006509 MBW18314.1 GALX01004399 JAB64067.1 MG993326 AYU58903.1 KU188517 ANE06405.1 KF792064 AHH25009.1 GDKW01000574 JAI56021.1 KT369806 AML25535.1 AY737550 AAU84943.1 KQ971319 EFA00468.1 GECL01003587 JAP02537.1 ABLF02032889 GEMB01002235 JAS00945.1 AY588061 AAT01073.1 NEVH01003508 PNF40452.1 DQ450899 ABE27979.1 KK853207 KDR09838.1 KC161211 AGJ76539.1 KX108734 ANV22067.1 KJ507199 AHY22050.1 FX983744 BAX07243.1 KU196668 AMD16548.1 GEZM01048387 GEZM01048386 JAV76591.1 ACPB03021529 GAHY01000509 JAA77001.1 APGK01027120 KB740648 KB632292 ENN79784.1 ERL91608.1 KU521368 KX428475 ANS71248.1 ANW09586.1 GDIP01175047 GDIP01170376 JAJ53026.1 GECZ01027298 GECZ01011797 JAS42471.1 JAS57972.1 DS235250 EEB13950.1 GEDC01023461 GEDC01016965 GEDC01002583 JAS13837.1 JAS20333.1 JAS34715.1 GBHO01044314 GBRD01005390 JAF99289.1 JAG60431.1 HACA01022742 CDW40103.1
Proteomes
UP000037510
UP000218220
UP000053240
UP000076408
UP000005204
UP000007151
+ More
UP000075885 UP000075883 UP000075886 UP000075903 UP000075884 UP000075902 UP000075920 UP000002320 UP000008820 UP000030765 UP000075882 UP000075880 UP000076407 UP000007062 UP000069272 UP000075900 UP000075840 UP000069940 UP000249989 UP000092462 UP000078492 UP000078540 UP000002358 UP000078541 UP000005205 UP000075809 UP000192223 UP000053097 UP000007755 UP000079169 UP000007266 UP000007819 UP000235965 UP000027135 UP000015103 UP000019118 UP000030742 UP000009046
UP000075885 UP000075883 UP000075886 UP000075903 UP000075884 UP000075902 UP000075920 UP000002320 UP000008820 UP000030765 UP000075882 UP000075880 UP000076407 UP000007062 UP000069272 UP000075900 UP000075840 UP000069940 UP000249989 UP000092462 UP000078492 UP000078540 UP000002358 UP000078541 UP000005205 UP000075809 UP000192223 UP000053097 UP000007755 UP000079169 UP000007266 UP000007819 UP000235965 UP000027135 UP000015103 UP000019118 UP000030742 UP000009046
Pfam
PF00022 Actin
ProteinModelPortal
X5CT98
V9NEM2
A0A0L7LTL0
A0A2W1BR20
A0A2A4JIS6
I4DLT1
+ More
A0A0N1IAY6 A0A2Z5STE6 B2Z6M6 T1DFC0 U5EUS3 A0A1W6S692 A0A2H1X0F8 A0A182YFN1 H9JWN1 A0A291FJB0 A0A2Z4N445 A0A212EZ57 A0A182P6B7 A0A182M762 A0A182PZM2 A0A182V6G0 A0A182N891 A0A182U5H7 A0A182WQH5 B0XB41 Q17KG3 A0A084W2J0 A0A182LB97 A0A182ITR0 A0A182X497 A8B1J8 A0A182FZK9 A0JCK7 A0A182RTL3 A0A182HPA0 A0A023ES50 P49871 B2DBP0 B0LF73 A0A1B0D7H8 A0A151JMQ0 A0A151I4W0 K7J803 A0A195FX88 A0A158NXU2 A0A3G1SUZ3 A0A151WIN4 Q6ELZ7 Q45L89 D3KU63 A0A1W4XH28 A0A2Z4G633 A7WPD3 A0A0P5CGV3 A0A026X3P0 E2IV57 P07836 F4WLU8 A0A1W4XWB9 A0A2R4GMB1 F2D4P0 A0A224XRR1 Q5XXT4 Q3HNA6 Q5RLJ4 A0A125RA11 A0A2D1QW10 A0A2H8TVL9 V5GR51 A0A1S3CVP9 A0A3G4R8Q5 A0A3G1DKW0 A0A023J916 A0A0P4VVE9 A0A140EFS7 Q5XUA4 D6WF19 A0A0V0G386 X1X262 A0A170ZGA9 Q6PPI5 A0A2J7RHY5 A1BPP6 A0A067QL86 M9VDW4 A0A1S5RIQ2 A0A023VZM4 A0A2Z5RE76 A0A125RA09 A0A1Y1LUA6 R4FQA8 N6TH73 A0A1B1UZE0 A0A0P5CHV2 A0A1B6EXE0 E0VKP4 A0A1B6CK57 A0A0A9W354 A0A0K2UPC6
A0A0N1IAY6 A0A2Z5STE6 B2Z6M6 T1DFC0 U5EUS3 A0A1W6S692 A0A2H1X0F8 A0A182YFN1 H9JWN1 A0A291FJB0 A0A2Z4N445 A0A212EZ57 A0A182P6B7 A0A182M762 A0A182PZM2 A0A182V6G0 A0A182N891 A0A182U5H7 A0A182WQH5 B0XB41 Q17KG3 A0A084W2J0 A0A182LB97 A0A182ITR0 A0A182X497 A8B1J8 A0A182FZK9 A0JCK7 A0A182RTL3 A0A182HPA0 A0A023ES50 P49871 B2DBP0 B0LF73 A0A1B0D7H8 A0A151JMQ0 A0A151I4W0 K7J803 A0A195FX88 A0A158NXU2 A0A3G1SUZ3 A0A151WIN4 Q6ELZ7 Q45L89 D3KU63 A0A1W4XH28 A0A2Z4G633 A7WPD3 A0A0P5CGV3 A0A026X3P0 E2IV57 P07836 F4WLU8 A0A1W4XWB9 A0A2R4GMB1 F2D4P0 A0A224XRR1 Q5XXT4 Q3HNA6 Q5RLJ4 A0A125RA11 A0A2D1QW10 A0A2H8TVL9 V5GR51 A0A1S3CVP9 A0A3G4R8Q5 A0A3G1DKW0 A0A023J916 A0A0P4VVE9 A0A140EFS7 Q5XUA4 D6WF19 A0A0V0G386 X1X262 A0A170ZGA9 Q6PPI5 A0A2J7RHY5 A1BPP6 A0A067QL86 M9VDW4 A0A1S5RIQ2 A0A023VZM4 A0A2Z5RE76 A0A125RA09 A0A1Y1LUA6 R4FQA8 N6TH73 A0A1B1UZE0 A0A0P5CHV2 A0A1B6EXE0 E0VKP4 A0A1B6CK57 A0A0A9W354 A0A0K2UPC6
PDB
5CE3
E-value=0,
Score=2022
Ontologies
PATHWAY
PANTHER
Topology
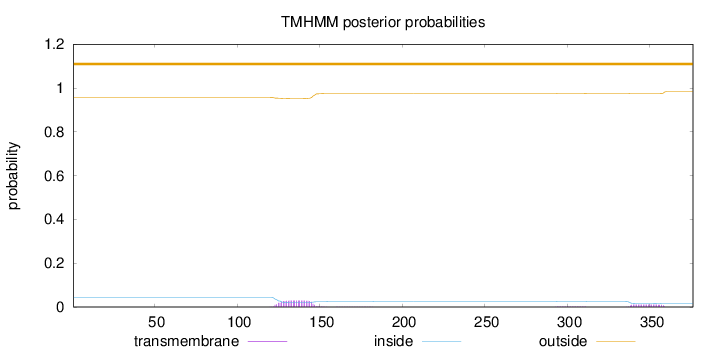
Subcellular location
Length:
376
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.893579999999999
Exp number, first 60 AAs:
0.00151
Total prob of N-in:
0.04341
outside
1 - 376
Population Genetic Test Statistics
Pi
202.336798
Theta
180.568102
Tajima's D
0.366746
CLR
20.259396
CSRT
0.471176441177941
Interpretation
Uncertain
