Gene
KWMTBOMO16148
Pre Gene Modal
BGIBMGA004523
Annotation
PREDICTED:_uncharacterized_protein_LOC101743883_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.694 PlasmaMembrane Reliability : 2.197
Sequence
CDS
ATGTACTCTAGTCACTCTGGTCTTCTACTGGAGAAATTCGCTTCTGATCCACTGCCGGGCAAGCATGTCCCGTTGGAGAGCTACGAAAGCGAGTTGGAGAGAGAGCACGCGCTGCCGACTTCCGTGCCCAACGCGGACATACTCAAGACTAACAGCAACCCCACGTACCACAAGCCCAGACCGCCCCAGCATCATGGAGCCTCCCTGCTGGCGGGCGTCGGGGCGGTGCCTTCGTACGCGCCTCCCGGACGAGCGTTCTTCACACCGCCTTTGCCACCGGAATACAAAAACCCATTTGCCGATAAGCCGACACTTAGAGGCACGAATACAGACGGGAACAATTATCTTAACAGGAGGCCCATACCTCCCCCCTCCCTGGGGCCGGGACACGAGAGGATACCGATCAGACCTCCGGGACAGGAGAGCTCTCCCCCGCAGGTGCCGGCGCCGCCCCCCGCGCCCCCGGCCGCACCTGACGTCACACACAGGAAGATACCGCTCAACACTCCTAGCCATAAGCCGGACGAACTAGAACCTGACGGTAAACAATCCCAAGCCAACTTTACGGACTTTGTTCCGAATTCACTCGACATTCCTGCCATAACTCGGATACTGTCCGGATCTAACGGCAGGAAAGAGGACATTCCGGATGTCCTGCTCAGGACAGTAACGACTAAACCTCAACACAGCCAGGAGAAGACCTCGAAAAGTGACATTTTTGTTACAAAAGACGTTGGCGTCACTCTCAGTGATACCAACAAAGACACATCCACTGAAGGATCAGTGTTACCAGTCCTAGATCCAGACATGAACAATCTAGACCGGCCGTTGATCATAGATCAAAACGGCGAAACTAATACCAGGAGAAACATACAATATTCTACTAAAAACGAAACCAAGAAAGAGTACACGCCAGGGACGACCGTAGGGTTCGAGCTAAAAGACCCCGAGATGTCCACTGAGAGGTACCACAACCTTGCCAACGTCAACTCGGACAAGAAGATCCTGAAGCAGGATCCGAAGGGCGGCGGCGCGGTGCAGCGACAGGGCGCGACCTGGCCCTTCGCCTGGAATGTGCACATCTATTTCGCGACGGTCATGTTCACGATATTGGCCGTCTACTCAATCTTTAAAATCATATGCTACAACAAGTTCACCCACCTCTTCACTCAATACTACTTCATCATACTCCATTTGATATTAGTTTTCGTTTGTTTGATGAGGATATTCTACATGTGCTACGATCCTTATAACATTAATAATTCACTACCGATTTTCATCAGTGAAATTTTGCTTCACATTCCCGAAATATTCTTGAGCATAGCATTCGCTTGTACCATCCTCTTCTTGCTCGTACACAGCATGAACTTCAAGAACAATTGCTGTTCGTTTCTGCTCCGGCCCAGGGTCATCATCGCGGGGGGCGGGGTCCACGTGCTGCTGTGCGTGATGTTGCACATACTCGAAACCAATAGCACCATATACAAAACGCAGAACTCAGAAAAGAGAATCTTAGGTCTGACCTGCGAAATTATCTTCATAATTGCGTGTCTAACTTTAGGTCTTGGTTATTTGTATGTATTCAAAATGCTTAAGACGGTCCTACAAAAGAAAAGTCACACGTACATAAATGGCTTCCAAAATCTATATCAGGCCATACACATAAATTTGGCGACTGCCTTGCTTTTTATACTAATGGCAACGTTACAAGTTTATGGTATATTTGGTATAAATCAAGTGAACATGAGTAAATTTAACTGGCTGCAGTGGGGCTACCAGTTCTCCTTGAGGCTGATAGAGATCAGCATAGTAGCGTTAATATCTTGGGTAATCAGTTTGAAGGTAGGCGTTGGTCCGCTTCAGCACGAAAAGGCTGACGGCAGCAAAATTCCAGGCTTGGGCCTTTTCACGTGCGGGGCTGGATCTAGTAATGAACAATTTGAGTCTGACTATCCAGCCATATGTAATACTAACACAAATCTTCATACCTACACGTTAAGAACTGGTAAACCGATATATGAAAGTGCCGGGCATCATCCGAACATGGAACCATGTTGTGGACCTATGGACCACCCAAGGTTCCTCAATACCTTGTCTGGTCCGTGTGATAACAATTCCGGTACTGAAAATTACTCCGGCCATAGTTCTATAGCCAACGCTGGTCACAGCAGTTCAACGGAATCTAGCAACGCGGCTTCCAGTCAGGGCGTGACCAACCATTTGATCAAACACCACCCAAATATGGGGTACAACGGCATGGAGTCTGCGTCCGACGACCACTACTCGAACTGCGACCCCGCAATACAGAGCTTGCAAGGAGACGACCCTTTGGATCATGAATACAACGTCATCCAATATGGTAGCAACAGTGACTTCATTCGAGATATCAAAAACCAGAACTACAACGGCGGCTCCAACAGGTCCTCGCACAACTTCAAGCACATGTCCTACGACAGGACGTCCTCTAGAACGAACAGCAATCCCCGGAAAAAGCACAGAGCGAAAAACAAAAACGTTCAAAACTGCATGACCATGGGCTACGACTCCGCCGCCGGCCACCACTACCACCAGCCGCACGTGCCGGACGAGTACGGCAACTACAACACAGAGAACGCCTCCTACCACACGTCCGGAATACAGACCCTCAACCCTAACAGGAACTACAGCAACGAATGTCCGGTAAGGACCTCGCAGCGTCGGCACTCTCCATCGACCTCCATGGTCAATTCGAGTGGTAGCCAAAAACGTGGTAAGGCGAATCCATTCCCGGACAGACCGAAGTCGACTGATCTATACGGGTTCGAAGAGCCCGGGAAGTATCCTCACGGGATGGGGACGCCCCAGCCGGCTCCGCGGAACTGTAGCTATGAGGTCTCGTATTCCCAGCCTCAAGACGAGATCCCCAACGAGGCGGGCAGCAGCATGCTGGTGGCGCAAGATGGCTTCGTCAGGTTCAGGTCGCTGGAAGACGGACCCTCGCAAACCTGA
Protein
MYSSHSGLLLEKFASDPLPGKHVPLESYESELEREHALPTSVPNADILKTNSNPTYHKPRPPQHHGASLLAGVGAVPSYAPPGRAFFTPPLPPEYKNPFADKPTLRGTNTDGNNYLNRRPIPPPSLGPGHERIPIRPPGQESSPPQVPAPPPAPPAAPDVTHRKIPLNTPSHKPDELEPDGKQSQANFTDFVPNSLDIPAITRILSGSNGRKEDIPDVLLRTVTTKPQHSQEKTSKSDIFVTKDVGVTLSDTNKDTSTEGSVLPVLDPDMNNLDRPLIIDQNGETNTRRNIQYSTKNETKKEYTPGTTVGFELKDPEMSTERYHNLANVNSDKKILKQDPKGGGAVQRQGATWPFAWNVHIYFATVMFTILAVYSIFKIICYNKFTHLFTQYYFIILHLILVFVCLMRIFYMCYDPYNINNSLPIFISEILLHIPEIFLSIAFACTILFLLVHSMNFKNNCCSFLLRPRVIIAGGGVHVLLCVMLHILETNSTIYKTQNSEKRILGLTCEIIFIIACLTLGLGYLYVFKMLKTVLQKKSHTYINGFQNLYQAIHINLATALLFILMATLQVYGIFGINQVNMSKFNWLQWGYQFSLRLIEISIVALISWVISLKVGVGPLQHEKADGSKIPGLGLFTCGAGSSNEQFESDYPAICNTNTNLHTYTLRTGKPIYESAGHHPNMEPCCGPMDHPRFLNTLSGPCDNNSGTENYSGHSSIANAGHSSSTESSNAASSQGVTNHLIKHHPNMGYNGMESASDDHYSNCDPAIQSLQGDDPLDHEYNVIQYGSNSDFIRDIKNQNYNGGSNRSSHNFKHMSYDRTSSRTNSNPRKKHRAKNKNVQNCMTMGYDSAAGHHYHQPHVPDEYGNYNTENASYHTSGIQTLNPNRNYSNECPVRTSQRRHSPSTSMVNSSGSQKRGKANPFPDRPKSTDLYGFEEPGKYPHGMGTPQPAPRNCSYEVSYSQPQDEIPNEAGSSMLVAQDGFVRFRSLEDGPSQT
Summary
Uniprot
Pubmed
EMBL
BABH01035463
NWSH01000655
PCG74995.1
RSAL01000078
RVE48724.1
ODYU01010038
+ More
SOQ54860.1 AGBW02009632 OWR50278.1 JTDY01001370 KOB74049.1 GDQN01004882 JAT86172.1 UFQS01000873 UFQT01000873 SSX07416.1 SSX27758.1 CH902633 KPU74653.1 CH479221 EDW34854.1 CH475449 EAL29288.2 KRT05306.1 CP012527 ALC48107.1 CH480867 EDW53718.1 CM002916 KMZ07162.1 KMZ07163.1 AE014135 AFH06770.1 AAF59363.2 CH933813 EDW10884.2 KRG07216.1 KRG07217.1 KRG07218.1 OUUW01000017 SPP89051.1 SPP89052.1 CM000161 EDW99295.1 SPP89053.1 CH954184 EDV45265.2 KB631749 ERL85784.1 LJIG01016055 KRT81750.1
SOQ54860.1 AGBW02009632 OWR50278.1 JTDY01001370 KOB74049.1 GDQN01004882 JAT86172.1 UFQS01000873 UFQT01000873 SSX07416.1 SSX27758.1 CH902633 KPU74653.1 CH479221 EDW34854.1 CH475449 EAL29288.2 KRT05306.1 CP012527 ALC48107.1 CH480867 EDW53718.1 CM002916 KMZ07162.1 KMZ07163.1 AE014135 AFH06770.1 AAF59363.2 CH933813 EDW10884.2 KRG07216.1 KRG07217.1 KRG07218.1 OUUW01000017 SPP89051.1 SPP89052.1 CM000161 EDW99295.1 SPP89053.1 CH954184 EDV45265.2 KB631749 ERL85784.1 LJIG01016055 KRT81750.1
Proteomes
Interpro
IPR023298
ATPase_P-typ_TM_dom_sf
SUPFAM
SSF81665
SSF81665
ProteinModelPortal
Ontologies
GO
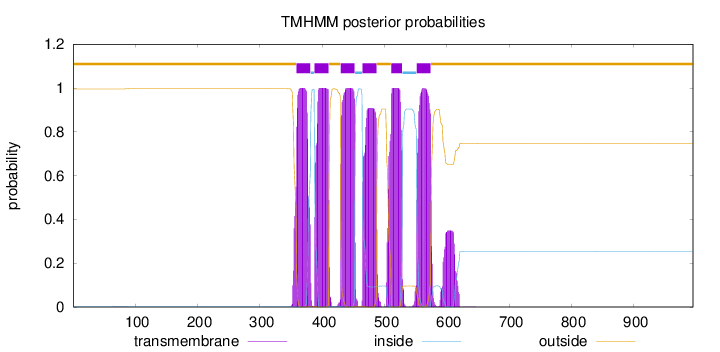
Topology
Length:
995
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
139.83415
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00383
outside
1 - 358
TMhelix
359 - 381
inside
382 - 387
TMhelix
388 - 410
outside
411 - 429
TMhelix
430 - 452
inside
453 - 464
TMhelix
465 - 487
outside
488 - 510
TMhelix
511 - 528
inside
529 - 551
TMhelix
552 - 574
outside
575 - 995
Population Genetic Test Statistics
Pi
244.950279
Theta
171.907752
Tajima's D
1.444253
CLR
0.080898
CSRT
0.773861306934653
Interpretation
Uncertain
