Gene
KWMTBOMO16145
Pre Gene Modal
BGIBMGA004525
Annotation
PREDICTED:_sugar_transporter_protein_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.942
Sequence
CDS
ATGAGGGTCACACCTTTTGTGAAGCAGTGCTTTGCGGTATCGAGTGTCGTCCTGAACATGATGTCGTATGGCCTTGTGGCCGGATTCAACGCTTCCCTGTTCTCTGAACTCAGGAGGACCCGAGAGATACCTCTGGACATACATTCAGAGTCTTGGTTGGCATCTTTGATCGGATTAACGCTTTTTGGGGGCTCTGTGATTAGTTCACTAGTGATGGAAGCCATTGGACGGAGACCAGCTATTCTAGCGAGTTCAGTTTTTATGATTTGCGGGTGGATATTGCTCAGTCTAGCTTCATCCTTTCCTGTCTTGTTAACGGCGAAAATTTGTCATGGAATATCCTTTGGATTAGAATCACCGAGCTTTCTGGTAACTAAGGGACGTTACGATGAATGCAGGAAAGTTTTCCGCTGGTTGAGGGGAACAGATGAGAATGACGAACTCGAAACAATGATTAAAACTCATATGGTACTTAAAGCAAATAAGCAAAGGCAGAGAATTAAAGTATTAACATTAATCAAGAACAAAGCGACATGCATCGTGACAGCTTTTAAAAAGAGAGAAGTTCGAATTCCAATAATTATTTCGATGCACTTGATGGCTATAAATCAGTTCTGCGGGTCTACGGTTAACGACATGTATGCTCTAGACGTCCACACCGCGTTGTACGGAACTAATGTCTACATGTTCAAAGTAATGACGTCACTAGATGTGCTAAGACTCACTGCGGCTCTGTCAGCAGTTTATATAACAAAAAAATTAGGGCGGCGTTCCATGCTGCTAATATTTGTGTCCTTGAATATATTCGTCAACCTGTGCCTAGCTGGACACATTTTTGCAAGACAACGTGATCTTCTACCTGCTTCACATATGGTAATAGGTGTCATTCTGCATCATTTTCAATGCTTCATTATGGGGACTGGATCCATACCCTTGATATTTGTAATTGCTGGTGAAATATTCCCTTTGGAATATAGGGGTTTATGTTGGATGGTGTCAAGTATATTCAGCTCCACTTTCATGTTCGTGAACATCAAATCAGCTCCGTATCTGTTCTCGTTGTTCGGCGTTGACGGGACGTTATGTTTTTATGCTTTGATTCTTGCCTATAGTCTAGTTGTCACTCGTATTTATTTACCGGAGACTAAAGACAAAACTCTTCAGGAAGTCGAAGATGAAATTAGGGGATTCGCTCTCCTCAGTCACGAAGAAGCGTTTGAGTTGAAGAGTGATAAAGCGGATGGGGAAGATTAA
Protein
MRVTPFVKQCFAVSSVVLNMMSYGLVAGFNASLFSELRRTREIPLDIHSESWLASLIGLTLFGGSVISSLVMEAIGRRPAILASSVFMICGWILLSLASSFPVLLTAKICHGISFGLESPSFLVTKGRYDECRKVFRWLRGTDENDELETMIKTHMVLKANKQRQRIKVLTLIKNKATCIVTAFKKREVRIPIIISMHLMAINQFCGSTVNDMYALDVHTALYGTNVYMFKVMTSLDVLRLTAALSAVYITKKLGRRSMLLIFVSLNIFVNLCLAGHIFARQRDLLPASHMVIGVILHHFQCFIMGTGSIPLIFVIAGEIFPLEYRGLCWMVSSIFSSTFMFVNIKSAPYLFSLFGVDGTLCFYALILAYSLVVTRIYLPETKDKTLQEVEDEIRGFALLSHEEAFELKSDKADGED
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
D0PWZ5
H9J4T5
E0X905
H9J4T7
H9J4U8
H9J4R7
+ More
H9J4S0 A0A2H1WVP2 A0A2W1BG84 A0A2A4KAY2 A0A3S2P2D1 H9J4R6 A0A023PNK9 A0A212F161 A0A2A4JTG8 A0A0N1IHN6 A0A194QPY2 A0A0L7L8D8 A0A2H1W936 A0A212FJ74 A0A2A4JUU3 A0A2W1BD78 A0A194QJI7 A0A437ATT4 H9J4S1 H9J4X0 A0A437BK01 A0A2W1BBH7 H9J4X1 A0A2H1V0I3 A0A0N1IPS2 A0A2A4KA42 A0A0N1I9K7 A0A437BR67 H9J4X7 A0A2W1B9E8 A0A2A4JU29 A0A194QJ47 A0A2A4JSU4 A0A0L7L8F3 A0A2A4JT91 H9J4X6 A0A437BJF6 A0A2H1WFL6 A0A212FCC0 A0A2H1WAU6 A0A212FPF6 H9J525 A0A3S2LUZ6 A0A2H1WB04 H9J533 A0A2H1W9B8 A0A212F9B1 A0A2A4J748 A0A2W1BFI0 H9J4W9 A0A2H1WK46 A0A2A4JNG3 B0W5I2 A0A0L7LRT2 A0A212F9C9 A0A2H1WAF2 A0A194QJ40 A0A0N1IG19 A0A194QJ30 A0A3S2LEF2 A0A194QJJ6 A0A182GCQ0 B0W5I1 A0A2H1X0D6 A0A182GTB3 Q17LS5 A0A0N0PAH4 A0A0N0PEE0 A0A2W1BMU2 A0A0N0PEP7 H9J017 H9JAI4
H9J4S0 A0A2H1WVP2 A0A2W1BG84 A0A2A4KAY2 A0A3S2P2D1 H9J4R6 A0A023PNK9 A0A212F161 A0A2A4JTG8 A0A0N1IHN6 A0A194QPY2 A0A0L7L8D8 A0A2H1W936 A0A212FJ74 A0A2A4JUU3 A0A2W1BD78 A0A194QJI7 A0A437ATT4 H9J4S1 H9J4X0 A0A437BK01 A0A2W1BBH7 H9J4X1 A0A2H1V0I3 A0A0N1IPS2 A0A2A4KA42 A0A0N1I9K7 A0A437BR67 H9J4X7 A0A2W1B9E8 A0A2A4JU29 A0A194QJ47 A0A2A4JSU4 A0A0L7L8F3 A0A2A4JT91 H9J4X6 A0A437BJF6 A0A2H1WFL6 A0A212FCC0 A0A2H1WAU6 A0A212FPF6 H9J525 A0A3S2LUZ6 A0A2H1WB04 H9J533 A0A2H1W9B8 A0A212F9B1 A0A2A4J748 A0A2W1BFI0 H9J4W9 A0A2H1WK46 A0A2A4JNG3 B0W5I2 A0A0L7LRT2 A0A212F9C9 A0A2H1WAF2 A0A194QJ40 A0A0N1IG19 A0A194QJ30 A0A3S2LEF2 A0A194QJJ6 A0A182GCQ0 B0W5I1 A0A2H1X0D6 A0A182GTB3 Q17LS5 A0A0N0PAH4 A0A0N0PEE0 A0A2W1BMU2 A0A0N0PEP7 H9J017 H9JAI4
EMBL
FJ373021
ACQ66002.1
BABH01035466
BABH01035467
GQ871755
ADM86146.1
+ More
BABH01035469 BABH01035543 BABH01035544 BABH01035538 BABH01035539 ODYU01011356 SOQ57052.1 KZ150249 PZC71836.1 NWSH01000008 PCG80953.1 RSAL01000045 RVE50592.1 BABH01035548 KJ439227 KZ150491 AHX25878.1 PZC70688.1 AGBW02010956 OWR47467.1 NWSH01000704 PCG74682.1 KQ460077 KPJ17887.1 KQ458671 KPJ05561.1 JTDY01002352 KOB71610.1 ODYU01007122 SOQ49615.1 AGBW02008317 OWR53759.1 NWSH01000578 PCG75536.1 PZC71835.1 KPJ05554.1 RSAL01000460 RVE41642.1 BABH01035527 BABH01035528 BABH01035529 BABH01025733 RVE50588.1 PZC70687.1 BABH01025731 ODYU01000118 SOQ34347.1 KPJ17881.1 PCG80949.1 KPJ17880.1 RSAL01000017 RVE52904.1 BABH01025717 PZC71838.1 PCG75537.1 KPJ05583.1 PCG74684.1 KOB71609.1 PCG74683.1 BABH01025720 RVE50594.1 ODYU01008354 SOQ51875.1 AGBW02009205 OWR51384.1 SOQ49614.1 AGBW02002687 OWR55626.1 BABH01038881 RSAL01000181 RVE44925.1 ODYU01007347 SOQ50032.1 BABH01038850 SOQ49613.1 AGBW02009614 OWR50324.1 NWSH01002771 PCG67556.1 KZ150133 PZC73021.1 ODYU01009098 SOQ53272.1 NWSH01001042 PCG72950.1 DS231843 EDS35465.1 JTDY01000230 KOB78170.1 OWR50323.1 SOQ50033.1 KPJ05573.1 KPJ17891.1 KPJ05563.1 RVE44926.1 KPJ05559.1 JXUM01009472 KQ560284 KXJ83288.1 EDS35464.1 ODYU01012470 SOQ58795.1 JXUM01086634 KQ563580 KXJ73638.1 CH477213 EAT47626.1 KQ458842 KPJ04897.1 KQ459890 KPJ19576.1 KZ150062 PZC74196.1 KPJ19577.1 BABH01018801 BABH01009421
BABH01035469 BABH01035543 BABH01035544 BABH01035538 BABH01035539 ODYU01011356 SOQ57052.1 KZ150249 PZC71836.1 NWSH01000008 PCG80953.1 RSAL01000045 RVE50592.1 BABH01035548 KJ439227 KZ150491 AHX25878.1 PZC70688.1 AGBW02010956 OWR47467.1 NWSH01000704 PCG74682.1 KQ460077 KPJ17887.1 KQ458671 KPJ05561.1 JTDY01002352 KOB71610.1 ODYU01007122 SOQ49615.1 AGBW02008317 OWR53759.1 NWSH01000578 PCG75536.1 PZC71835.1 KPJ05554.1 RSAL01000460 RVE41642.1 BABH01035527 BABH01035528 BABH01035529 BABH01025733 RVE50588.1 PZC70687.1 BABH01025731 ODYU01000118 SOQ34347.1 KPJ17881.1 PCG80949.1 KPJ17880.1 RSAL01000017 RVE52904.1 BABH01025717 PZC71838.1 PCG75537.1 KPJ05583.1 PCG74684.1 KOB71609.1 PCG74683.1 BABH01025720 RVE50594.1 ODYU01008354 SOQ51875.1 AGBW02009205 OWR51384.1 SOQ49614.1 AGBW02002687 OWR55626.1 BABH01038881 RSAL01000181 RVE44925.1 ODYU01007347 SOQ50032.1 BABH01038850 SOQ49613.1 AGBW02009614 OWR50324.1 NWSH01002771 PCG67556.1 KZ150133 PZC73021.1 ODYU01009098 SOQ53272.1 NWSH01001042 PCG72950.1 DS231843 EDS35465.1 JTDY01000230 KOB78170.1 OWR50323.1 SOQ50033.1 KPJ05573.1 KPJ17891.1 KPJ05563.1 RVE44926.1 KPJ05559.1 JXUM01009472 KQ560284 KXJ83288.1 EDS35464.1 ODYU01012470 SOQ58795.1 JXUM01086634 KQ563580 KXJ73638.1 CH477213 EAT47626.1 KQ458842 KPJ04897.1 KQ459890 KPJ19576.1 KZ150062 PZC74196.1 KPJ19577.1 BABH01018801 BABH01009421
Proteomes
PRIDE
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
D0PWZ5
H9J4T5
E0X905
H9J4T7
H9J4U8
H9J4R7
+ More
H9J4S0 A0A2H1WVP2 A0A2W1BG84 A0A2A4KAY2 A0A3S2P2D1 H9J4R6 A0A023PNK9 A0A212F161 A0A2A4JTG8 A0A0N1IHN6 A0A194QPY2 A0A0L7L8D8 A0A2H1W936 A0A212FJ74 A0A2A4JUU3 A0A2W1BD78 A0A194QJI7 A0A437ATT4 H9J4S1 H9J4X0 A0A437BK01 A0A2W1BBH7 H9J4X1 A0A2H1V0I3 A0A0N1IPS2 A0A2A4KA42 A0A0N1I9K7 A0A437BR67 H9J4X7 A0A2W1B9E8 A0A2A4JU29 A0A194QJ47 A0A2A4JSU4 A0A0L7L8F3 A0A2A4JT91 H9J4X6 A0A437BJF6 A0A2H1WFL6 A0A212FCC0 A0A2H1WAU6 A0A212FPF6 H9J525 A0A3S2LUZ6 A0A2H1WB04 H9J533 A0A2H1W9B8 A0A212F9B1 A0A2A4J748 A0A2W1BFI0 H9J4W9 A0A2H1WK46 A0A2A4JNG3 B0W5I2 A0A0L7LRT2 A0A212F9C9 A0A2H1WAF2 A0A194QJ40 A0A0N1IG19 A0A194QJ30 A0A3S2LEF2 A0A194QJJ6 A0A182GCQ0 B0W5I1 A0A2H1X0D6 A0A182GTB3 Q17LS5 A0A0N0PAH4 A0A0N0PEE0 A0A2W1BMU2 A0A0N0PEP7 H9J017 H9JAI4
H9J4S0 A0A2H1WVP2 A0A2W1BG84 A0A2A4KAY2 A0A3S2P2D1 H9J4R6 A0A023PNK9 A0A212F161 A0A2A4JTG8 A0A0N1IHN6 A0A194QPY2 A0A0L7L8D8 A0A2H1W936 A0A212FJ74 A0A2A4JUU3 A0A2W1BD78 A0A194QJI7 A0A437ATT4 H9J4S1 H9J4X0 A0A437BK01 A0A2W1BBH7 H9J4X1 A0A2H1V0I3 A0A0N1IPS2 A0A2A4KA42 A0A0N1I9K7 A0A437BR67 H9J4X7 A0A2W1B9E8 A0A2A4JU29 A0A194QJ47 A0A2A4JSU4 A0A0L7L8F3 A0A2A4JT91 H9J4X6 A0A437BJF6 A0A2H1WFL6 A0A212FCC0 A0A2H1WAU6 A0A212FPF6 H9J525 A0A3S2LUZ6 A0A2H1WB04 H9J533 A0A2H1W9B8 A0A212F9B1 A0A2A4J748 A0A2W1BFI0 H9J4W9 A0A2H1WK46 A0A2A4JNG3 B0W5I2 A0A0L7LRT2 A0A212F9C9 A0A2H1WAF2 A0A194QJ40 A0A0N1IG19 A0A194QJ30 A0A3S2LEF2 A0A194QJJ6 A0A182GCQ0 B0W5I1 A0A2H1X0D6 A0A182GTB3 Q17LS5 A0A0N0PAH4 A0A0N0PEE0 A0A2W1BMU2 A0A0N0PEP7 H9J017 H9JAI4
PDB
6N3I
E-value=1.91189e-06,
Score=124
Ontologies
GO
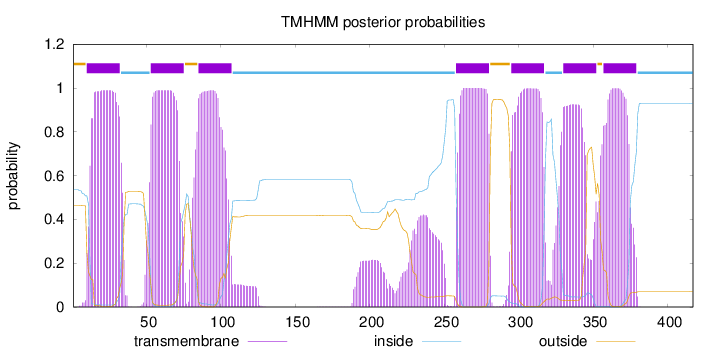
Topology
Length:
417
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
168.19139
Exp number, first 60 AAs:
31.24269
Total prob of N-in:
0.53624
POSSIBLE N-term signal
sequence
outside
1 - 9
TMhelix
10 - 32
inside
33 - 52
TMhelix
53 - 75
outside
76 - 84
TMhelix
85 - 107
inside
108 - 257
TMhelix
258 - 280
outside
281 - 294
TMhelix
295 - 317
inside
318 - 329
TMhelix
330 - 352
outside
353 - 356
TMhelix
357 - 379
inside
380 - 417
Population Genetic Test Statistics
Pi
237.677078
Theta
187.129473
Tajima's D
1.277654
CLR
0.195052
CSRT
0.732213389330533
Interpretation
Uncertain
