Gene
KWMTBOMO16134
Annotation
PREDICTED:_RNA-directed_DNA_polymerase_from_mobile_element_jockey-like_[Amyelois_transitella]
Full name
Guanylate cyclase
Location in the cell
PlasmaMembrane Reliability : 1.656
Sequence
CDS
ATGTCACGGGGTAAGTCTCCGGGTCATGACGAGCTAAGCATAGAACACCTAAAGTATGCCGGTGTACACTTGCCCCGAGTCTTGGTTATGTTCTACAATTTATGTATAGGACACTCCTACTTGCCTGAGGAGATGTTGAAGACTGTGGTAGTGCCAATTGTGAAGAATAGGACCGGGGATATATGTGATAAAACCAATTACAGACCCATCTCGTTAGCAACTATCATAGCGAAAGTGCTTGACAGTTTGCTTAACATACAACTTGATAAATATCTTCAGATACATGATAACCAGTTTGGTTTTAAATCAGGAATGTCTACAGAAATAGCCGTATTGTGTTTAAAGCAAACTGTCAAGTACTATACTGATCGAAAAACTCCAATTTTTGCCTGTTTCCTCGATTTATCAAAGGCATTCGACTTGGTTTGTTATGACATCTTATGGCAAAAACTGAATAATACTAAAATGCCGAAAGAACTAGTACAGATATTCAAATATTGGTACGCCAACCAGCTGAACGTGGTACGATGGTCGGGTGTCTATTCGAGGCCATACGGGATGGAGTGCGGGGTGAGGCAAGGAGGGTTGACCTCGCCCAAGCTCTTCAACCTGTATGTGAATGCTCTAATCGAGGAGCTCAGCAGTACTCATGTCGGCTGCCATATTGATGAAATATGTCTCAACAATATCAGTTATGCTGATGACATGGTGCTGCTGAGTGCGTCGGCCTGTGGGCTGTCTAAGCTGCTTGGCATATGTGAAAGGTATGCTCTAAGTCACGGTCTAGTGTATAATACAAAGAAAAGTGAGTGTATGGTGTTTCAGGCCAGAGGCAGGTGTCTGGATAACATTCCCTCCGTCAAACTGAATGGTACGCCACTGCGCAGAGTAGACAAGTTTAAATACCTAGGACACTTTGTAACCTCTGACCTGAGGGATGATACAGACATTGAGCGGGAGCGGAGAGCGCTGTCTGTAAGAGCTAATATGATTGCCCGCAGGTTCGCTCGGTGTTCGCGGGATGTCAAAATCACTCTATTTAGAGCGTTTTGTACGTCCTTCTATACGTGCAGCCTGTGGGTGTTCTATAGTCAAAAGCAGTACAGCGCCTTACGCGTCCAATACAACAATGCGTTCAGGGTGTTGTTGGGGCTGCCTCGGTTCTGTAGCGCGTCGGGAATGTTCGCCGAGGCACGAGTCGACTGTTTCTTTACGACCATGCGGAAAAGATGCACATCCCTGATTGGCCGGGTCCGGGTCAGCCCTAACAACATCCTGAAGGCATTAACAGACCGTTATGACTGTCGGTACTTGAACCATTGTACCATGGTTCACTTGCAGGCTAAATAA
Protein
MSRGKSPGHDELSIEHLKYAGVHLPRVLVMFYNLCIGHSYLPEEMLKTVVVPIVKNRTGDICDKTNYRPISLATIIAKVLDSLLNIQLDKYLQIHDNQFGFKSGMSTEIAVLCLKQTVKYYTDRKTPIFACFLDLSKAFDLVCYDILWQKLNNTKMPKELVQIFKYWYANQLNVVRWSGVYSRPYGMECGVRQGGLTSPKLFNLYVNALIEELSSTHVGCHIDEICLNNISYADDMVLLSASACGLSKLLGICERYALSHGLVYNTKKSECMVFQARGRCLDNIPSVKLNGTPLRRVDKFKYLGHFVTSDLRDDTDIERERRALSVRANMIARRFARCSRDVKITLFRAFCTSFYTCSLWVFYSQKQYSALRVQYNNAFRVLLGLPRFCSASGMFAEARVDCFFTTMRKRCTSLIGRVRVSPNNILKALTDRYDCRYLNHCTMVHLQAK
Summary
Catalytic Activity
GTP = 3',5'-cyclic GMP + diphosphate
Uniprot
A0A2W1BGX6
A0A2A4KA90
A0A2W1BJX4
A0A2W1BUS0
A0A2W1CP14
A0A3S2PB24
+ More
A0A2W1BE63 A0A3S2NHF1 S4NNA8 A0A2H1W8M9 A0A2W1BC01 A0A1A8UAJ7 A0A3B3HNF2 A0A3B3I5I8 A0A3B3IJ19 A0A3P9KQQ7 A0A3B3I0P1 A0A3P8PZH5 A0A146RF43 A0A146RB37 A0A146R782 A0A146ZPI4 A0A3P8RHR9 A0A146P365 A0A146RD82 A0A3C1S3F5 A0A0P4VXS2 A0A3C1S1R4 W4YPF0 A0A1Y1M3I5 A0A1Y1M3Z3 A0A3C1S364 A0A1Y1K7T4 A0A1Y1LZ08 D5LB38 A0A2W1BNH4 D7F163 A0A016VCK8 D7F178 A0A3M7SXU2 W4Y5D2 A0A3P9JDD8 A0A1Y1K5U2 A0A3M7Q8J1 A0A3B3I759 A0A1Y1K319 A0A0N7ZBB2 A0A3B3HTA8 A0A1T2KXF3 A0A0A9VZM2 A0A146KQ35 A0A3B1JC70 A0A0A9XQM2 A0A016SX67 D7F168 D7F175 D7F167 A0A3P9JDN6 A0A016TNH6 A0A016TNL1
A0A2W1BE63 A0A3S2NHF1 S4NNA8 A0A2H1W8M9 A0A2W1BC01 A0A1A8UAJ7 A0A3B3HNF2 A0A3B3I5I8 A0A3B3IJ19 A0A3P9KQQ7 A0A3B3I0P1 A0A3P8PZH5 A0A146RF43 A0A146RB37 A0A146R782 A0A146ZPI4 A0A3P8RHR9 A0A146P365 A0A146RD82 A0A3C1S3F5 A0A0P4VXS2 A0A3C1S1R4 W4YPF0 A0A1Y1M3I5 A0A1Y1M3Z3 A0A3C1S364 A0A1Y1K7T4 A0A1Y1LZ08 D5LB38 A0A2W1BNH4 D7F163 A0A016VCK8 D7F178 A0A3M7SXU2 W4Y5D2 A0A3P9JDD8 A0A1Y1K5U2 A0A3M7Q8J1 A0A3B3I759 A0A1Y1K319 A0A0N7ZBB2 A0A3B3HTA8 A0A1T2KXF3 A0A0A9VZM2 A0A146KQ35 A0A3B1JC70 A0A0A9XQM2 A0A016SX67 D7F168 D7F175 D7F167 A0A3P9JDN6 A0A016TNH6 A0A016TNL1
EC Number
4.6.1.2
Pubmed
EMBL
KZ150185
PZC72467.1
NWSH01000012
PCG80896.1
KZ150039
PZC74591.1
+ More
KZ149940 PZC76937.1 NHMN01024349 PZC87407.1 RSAL01000132 RVE46368.1 KZ150122 PZC73158.1 RSAL01004459 RVE40147.1 GAIX01014011 JAA78549.1 ODYU01007038 SOQ49431.1 KZ150625 PZC70440.1 HAEJ01003895 SBS44352.1 GCES01119762 JAQ66560.1 GCES01120657 JAQ65665.1 GCES01122004 JAQ64318.1 GCES01018090 JAR68233.1 GCES01147931 JAQ38391.1 GCES01120011 JAQ66311.1 DMQF01000507 HAO15811.1 GDRN01099830 JAI58698.1 DMQF01000358 HAO15222.1 AAGJ04127121 AAGJ04127122 AAGJ04127123 GEZM01043747 JAV78865.1 GEZM01043993 JAV78596.1 DMQF01000476 HAO15703.1 GEZM01094272 GEZM01094271 JAV55745.1 GEZM01043748 JAV78864.1 GU815089 ADF18552.1 KZ150025 PZC74827.1 FJ265548 ADI61816.1 JARK01001348 EYC25145.1 FJ265563 ADI61831.1 REGN01000623 RNA40614.1 AAGJ04058037 GEZM01094174 JAV55841.1 REGN01007025 RNA07484.1 GEZM01094173 JAV55843.1 GDRN01086563 JAI61160.1 MPRK01000277 OOZ37539.1 GBHO01042545 GBRD01013465 JAG01059.1 JAG52361.1 GDHC01021277 JAP97351.1 GBHO01021345 JAG22259.1 JARK01001500 JARK01001343 EYB95040.1 EYC28825.1 FJ265553 ADI61821.1 FJ265560 ADI61828.1 FJ265552 ADI61820.1 JARK01001424 EYC04315.1 EYC04316.1
KZ149940 PZC76937.1 NHMN01024349 PZC87407.1 RSAL01000132 RVE46368.1 KZ150122 PZC73158.1 RSAL01004459 RVE40147.1 GAIX01014011 JAA78549.1 ODYU01007038 SOQ49431.1 KZ150625 PZC70440.1 HAEJ01003895 SBS44352.1 GCES01119762 JAQ66560.1 GCES01120657 JAQ65665.1 GCES01122004 JAQ64318.1 GCES01018090 JAR68233.1 GCES01147931 JAQ38391.1 GCES01120011 JAQ66311.1 DMQF01000507 HAO15811.1 GDRN01099830 JAI58698.1 DMQF01000358 HAO15222.1 AAGJ04127121 AAGJ04127122 AAGJ04127123 GEZM01043747 JAV78865.1 GEZM01043993 JAV78596.1 DMQF01000476 HAO15703.1 GEZM01094272 GEZM01094271 JAV55745.1 GEZM01043748 JAV78864.1 GU815089 ADF18552.1 KZ150025 PZC74827.1 FJ265548 ADI61816.1 JARK01001348 EYC25145.1 FJ265563 ADI61831.1 REGN01000623 RNA40614.1 AAGJ04058037 GEZM01094174 JAV55841.1 REGN01007025 RNA07484.1 GEZM01094173 JAV55843.1 GDRN01086563 JAI61160.1 MPRK01000277 OOZ37539.1 GBHO01042545 GBRD01013465 JAG01059.1 JAG52361.1 GDHC01021277 JAP97351.1 GBHO01021345 JAG22259.1 JARK01001500 JARK01001343 EYB95040.1 EYC28825.1 FJ265553 ADI61821.1 FJ265560 ADI61828.1 FJ265552 ADI61820.1 JARK01001424 EYC04315.1 EYC04316.1
Proteomes
Pfam
Interpro
IPR036691
Endo/exonu/phosph_ase_sf
+ More
IPR000477 RT_dom
IPR024989 MFS_assoc_dom
IPR005135 Endo/exonuclease/phosphatase
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR013087 Znf_C2H2_type
IPR002138 Pept_C14_p10
IPR001828 ANF_lig-bd_rcpt
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR011009 Kinase-like_dom_sf
IPR001170 ANPR/GUC
IPR001054 A/G_cyclase
IPR028082 Peripla_BP_I
IPR000719 Prot_kinase_dom
IPR029787 Nucleotide_cyclase
IPR000477 RT_dom
IPR024989 MFS_assoc_dom
IPR005135 Endo/exonuclease/phosphatase
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR013087 Znf_C2H2_type
IPR002138 Pept_C14_p10
IPR001828 ANF_lig-bd_rcpt
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR011009 Kinase-like_dom_sf
IPR001170 ANPR/GUC
IPR001054 A/G_cyclase
IPR028082 Peripla_BP_I
IPR000719 Prot_kinase_dom
IPR029787 Nucleotide_cyclase
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A0A2W1BGX6
A0A2A4KA90
A0A2W1BJX4
A0A2W1BUS0
A0A2W1CP14
A0A3S2PB24
+ More
A0A2W1BE63 A0A3S2NHF1 S4NNA8 A0A2H1W8M9 A0A2W1BC01 A0A1A8UAJ7 A0A3B3HNF2 A0A3B3I5I8 A0A3B3IJ19 A0A3P9KQQ7 A0A3B3I0P1 A0A3P8PZH5 A0A146RF43 A0A146RB37 A0A146R782 A0A146ZPI4 A0A3P8RHR9 A0A146P365 A0A146RD82 A0A3C1S3F5 A0A0P4VXS2 A0A3C1S1R4 W4YPF0 A0A1Y1M3I5 A0A1Y1M3Z3 A0A3C1S364 A0A1Y1K7T4 A0A1Y1LZ08 D5LB38 A0A2W1BNH4 D7F163 A0A016VCK8 D7F178 A0A3M7SXU2 W4Y5D2 A0A3P9JDD8 A0A1Y1K5U2 A0A3M7Q8J1 A0A3B3I759 A0A1Y1K319 A0A0N7ZBB2 A0A3B3HTA8 A0A1T2KXF3 A0A0A9VZM2 A0A146KQ35 A0A3B1JC70 A0A0A9XQM2 A0A016SX67 D7F168 D7F175 D7F167 A0A3P9JDN6 A0A016TNH6 A0A016TNL1
A0A2W1BE63 A0A3S2NHF1 S4NNA8 A0A2H1W8M9 A0A2W1BC01 A0A1A8UAJ7 A0A3B3HNF2 A0A3B3I5I8 A0A3B3IJ19 A0A3P9KQQ7 A0A3B3I0P1 A0A3P8PZH5 A0A146RF43 A0A146RB37 A0A146R782 A0A146ZPI4 A0A3P8RHR9 A0A146P365 A0A146RD82 A0A3C1S3F5 A0A0P4VXS2 A0A3C1S1R4 W4YPF0 A0A1Y1M3I5 A0A1Y1M3Z3 A0A3C1S364 A0A1Y1K7T4 A0A1Y1LZ08 D5LB38 A0A2W1BNH4 D7F163 A0A016VCK8 D7F178 A0A3M7SXU2 W4Y5D2 A0A3P9JDD8 A0A1Y1K5U2 A0A3M7Q8J1 A0A3B3I759 A0A1Y1K319 A0A0N7ZBB2 A0A3B3HTA8 A0A1T2KXF3 A0A0A9VZM2 A0A146KQ35 A0A3B1JC70 A0A0A9XQM2 A0A016SX67 D7F168 D7F175 D7F167 A0A3P9JDN6 A0A016TNH6 A0A016TNL1
PDB
5HHL
E-value=0.000510793,
Score=103
Ontologies
GO
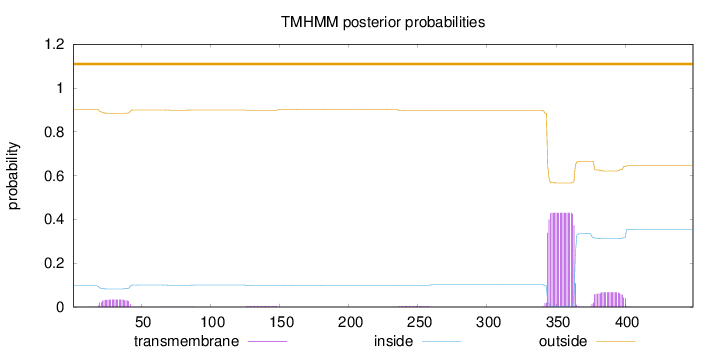
Topology
Length:
449
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
10.96234
Exp number, first 60 AAs:
0.70511
Total prob of N-in:
0.09721
outside
1 - 449
Population Genetic Test Statistics
Pi
156.240755
Theta
82.156061
Tajima's D
2.156058
CLR
0
CSRT
0.901654917254137
Interpretation
Uncertain
