Gene
KWMTBOMO16130 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004534
Annotation
PREDICTED:_plasma_membrane_calcium-transporting_ATPase_2_isoform_X7_[Bombyx_mori]
Full name
Calcium-transporting ATPase
Location in the cell
Cytoplasmic Reliability : 2.101
Sequence
CDS
ATGATTCACTGTCTTAGCGGTTCGAAGGCTGACCTGCAGCATCGGCGGGAAGTGTTCGGTTCAAACTTGATCCCGCCGAAGCCACCCAAGACTTTCCTGACGCTAGTATGGGAGGCTCTGCAGGATGTCACCCTTATTATCCTAGAAGTTGCTGCTGTTGTTTCATTAGGATTGAGTTTTTACAAGCCCGCAGATGACCCCAATGATGCGGCACATTTGGACGAAGAGGAGGGCCACTACCAGTGGATCGAGGGTCTGGCCATCTTAATATCGGTTATAGTCGTCGTCATAGTAACAGCGTTTAATGATTACACGAAAGAAAGGCAATTCAGAGGTCTGCAGTCTCGGATAGAAGGCGAGCACAAGTTCGCCGTGATCCGCGGCACGGAGGTCAAACAGGTGCCCATCAGCGAGATCGTCGTCGGCGACATATGTCAGATCAAGTACGGAGATCTGCTTCCGGCTGACGGTATCCTGCTGCAGAGCAACGATCTTAAGATTGACGAGTCATCCCTGACGGGAGAATCGGACCATGTCAAGAAGGGTGAAACATTTGATCCCATGGTGCTATCCGGTACACACGTTATGGAAGGTTCCGGCAAGATGTTAGTGACAGCCGTGGGGGTGAACTCCCAGGCCGGCATCATCTTCACGCTGCTCGGTGCCGCTGTCGACAAGCAGGAGAAGGAAATTAAACAGAAGAAGAAAGGTGAGGAGGAGGGAGTGGGGGGCGTGGGCGCCAACCACGCGCCGCCCGACGACAACCACGCGCCGCCCGCCGCCGACAAGCCGCCGCCAGAGCCGCAACACAAGAAGGAGAAGTCCGTGCTGCAGGCCAAACTTACCAAGCTCGCTATACAGATCGGTTACGCCGGCTCGACGATCGCCGTGCTCACGGTCATAATCCTGGTCATACAGTTCTGCGTCCGGACGTTCGCCATAGAGGGCAAGCCGTGGAAGGCGACCTACATTAACAACCTGGTGAAGCACCTCATCATCGGTGTGACGGTGCTGGTCGTCGCCGTGCCCGAGGGGCTGCCGCTCGCTGTCACACTCTCGCTTGCTTATTCCGTAAAGAAAATGATGAAAGACAACAATCTGGTGCGCCACCTGGACGCCTGCGAGACCATGGGCAACGCCACCGCCATCTGCTCGGACAAGACCGGCACGCTGACCACCAACCGCATGACGGTCGTGCAGTCGTACATCTGCGAGAAGCTGTGCAAGGTGACGCCCAACTACCGCGACATCCCGCCGCCCGTCGCCGACCTCATCGTGCAGGGCGTGGCGCTCAACTCCGCCTTCACCTCCAGGATCATGCCATCACAAGATCCAACGATGCCATCCATGCAAGTCGGTAACAAAACTGAATGCGCCCTCCTCGGATTCGTCCTGGCTCTAGGGCAGAGCTACGAAGCCGTCAGAGAAAGTCATCCCGAAGAGTCGTTCACCAGGGTGTACACCTTCAACTCGGTGCGGAAGAGCATGTCCACCGTCATACCGCACAACGGCGGCTACAGGCTCTACACTAAGGGAGCTTCGGAGATCGTGCTTAAAAAGTGCGCCTTCATCTACGGGCACGAGGGCCGCCTGGAGAAGTTCACGCGCGACATGCAGGAGCGGCTCGTGCGCCAGGTCATCGAGCCCATGGCCTGCGACGGGCTGCGGACCATCTCCGTGGCCTACCGGGACTTCGTGCCCGGGAAGGCGGACATCAACCAGGTGCACGTGGACCAGGAGCCGCACTGGGACGACGAGGACAACATCGTGAACAACCTGACCTGCCTCTGCGTTGTAGGGATCGAGGACCCCGTGAGGCCCGAGGTTCCAGAGGCCATCCGCAAGTGTCAGAAGGCCGGCATCACGGTGCGCATGGTGACGGGAGACAACGTGAACACCGCGCGCTCCATCGCCATGAAGTGCGGCATCCTCAGGCCCACGGACGACTTCCTGGTGCTCGAGGGGAAGGAGTTCAACCAACGCATACGGGACGCCAACGGAGAGGTGCAGCAGCAGCTGGTGGACAAGGTGTGGCCCAGCCTGCGCGTGCTGGCGCGCTCGTCGCCCACGGACAAGTACACGCTCGTCAAAGGCATGATCGAGTCCAAGGCATTCGACACGCGCGAGGTGGTGGCCGTCACCGGGGACGGGACCAACGACGGACCCGCGCTCAAGAAGGCCGACGTCGGATTCGCCATGGTGCGCCGCCACTACACGCACGCACACACGCACACACACACGCGCTGA
Protein
MIHCLSGSKADLQHRREVFGSNLIPPKPPKTFLTLVWEALQDVTLIILEVAAVVSLGLSFYKPADDPNDAAHLDEEEGHYQWIEGLAILISVIVVVIVTAFNDYTKERQFRGLQSRIEGEHKFAVIRGTEVKQVPISEIVVGDICQIKYGDLLPADGILLQSNDLKIDESSLTGESDHVKKGETFDPMVLSGTHVMEGSGKMLVTAVGVNSQAGIIFTLLGAAVDKQEKEIKQKKKGEEEGVGGVGANHAPPDDNHAPPAADKPPPEPQHKKEKSVLQAKLTKLAIQIGYAGSTIAVLTVIILVIQFCVRTFAIEGKPWKATYINNLVKHLIIGVTVLVVAVPEGLPLAVTLSLAYSVKKMMKDNNLVRHLDACETMGNATAICSDKTGTLTTNRMTVVQSYICEKLCKVTPNYRDIPPPVADLIVQGVALNSAFTSRIMPSQDPTMPSMQVGNKTECALLGFVLALGQSYEAVRESHPEESFTRVYTFNSVRKSMSTVIPHNGGYRLYTKGASEIVLKKCAFIYGHEGRLEKFTRDMQERLVRQVIEPMACDGLRTISVAYRDFVPGKADINQVHVDQEPHWDDEDNIVNNLTCLCVVGIEDPVRPEVPEAIRKCQKAGITVRMVTGDNVNTARSIAMKCGILRPTDDFLVLEGKEFNQRIRDANGEVQQQLVDKVWPSLRVLARSSPTDKYTLVKGMIESKAFDTREVVAVTGDGTNDGPALKKADVGFAMVRRHYTHAHTHTHTR
Summary
Description
This magnesium-dependent enzyme catalyzes the hydrolysis of ATP coupled with the transport of calcium.
Catalytic Activity
ATP + Ca(2+)(in) + H2O = ADP + Ca(2+)(out) + H(+) + phosphate
Similarity
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family.
Uniprot
A0A232F311
V9I6Y1
A0A0K8W474
A0A0K8U0F8
A0A0K8UCZ8
A0A034W2K7
+ More
A0A034W3M2 A0A034W478 A0A0K8V849 A0A034W033 A0A1W4VHY3 A0A0N8NZM1 A0A0P8XIK1 A0A1W4VIP7 A0A1I8P3E0 A0A1A9WYB0 T1P9V5 A0A1I8M346 A0A1I8PR34 A0A1I8MG26 A0A1S4ERF4 A0A3Q0IU77 B4PW03 A0A0R1ECC4 A0A0Q5SDQ7 E6EK15 A0A0Q5SQA8 A0A1B0F9W3 A0A1I8PK10 A0A0L7R0N1 A0A0M8ZY79 A0A310S5N1
A0A034W3M2 A0A034W478 A0A0K8V849 A0A034W033 A0A1W4VHY3 A0A0N8NZM1 A0A0P8XIK1 A0A1W4VIP7 A0A1I8P3E0 A0A1A9WYB0 T1P9V5 A0A1I8M346 A0A1I8PR34 A0A1I8MG26 A0A1S4ERF4 A0A3Q0IU77 B4PW03 A0A0R1ECC4 A0A0Q5SDQ7 E6EK15 A0A0Q5SQA8 A0A1B0F9W3 A0A1I8PK10 A0A0L7R0N1 A0A0M8ZY79 A0A310S5N1
EC Number
7.2.2.10
Pubmed
EMBL
NNAY01001107
OXU25111.1
JR035946
JR035949
AEY56812.1
AEY56815.1
+ More
GDHF01017233 GDHF01011200 GDHF01006296 JAI35081.1 JAI41114.1 JAI46018.1 GDHF01032117 GDHF01027167 JAI20197.1 JAI25147.1 GDHF01027762 GDHF01008159 GDHF01006384 JAI24552.1 JAI44155.1 JAI45930.1 GAKP01010025 GAKP01010023 JAC48929.1 GAKP01010032 GAKP01010030 JAC48922.1 GAKP01010026 GAKP01010022 JAC48930.1 GDHF01021214 GDHF01017197 JAI31100.1 JAI35117.1 GAKP01010031 GAKP01010029 JAC48921.1 CH902641 KPU74708.1 KPU74707.1 KA644945 AFP59574.1 CM000161 EDW99308.2 KRK04858.1 CH954184 KQS21513.1 AE014135 ADV37599.1 KQS21509.1 CCAG010020419 KQ414670 KOC64415.1 KQ435826 KOX72009.1 KQ769812 OAD52837.1
GDHF01017233 GDHF01011200 GDHF01006296 JAI35081.1 JAI41114.1 JAI46018.1 GDHF01032117 GDHF01027167 JAI20197.1 JAI25147.1 GDHF01027762 GDHF01008159 GDHF01006384 JAI24552.1 JAI44155.1 JAI45930.1 GAKP01010025 GAKP01010023 JAC48929.1 GAKP01010032 GAKP01010030 JAC48922.1 GAKP01010026 GAKP01010022 JAC48930.1 GDHF01021214 GDHF01017197 JAI31100.1 JAI35117.1 GAKP01010031 GAKP01010029 JAC48921.1 CH902641 KPU74708.1 KPU74707.1 KA644945 AFP59574.1 CM000161 EDW99308.2 KRK04858.1 CH954184 KQS21513.1 AE014135 ADV37599.1 KQS21509.1 CCAG010020419 KQ414670 KOC64415.1 KQ435826 KOX72009.1 KQ769812 OAD52837.1
Proteomes
PRIDE
Interpro
IPR008250
ATPase_P-typ_transduc_dom_A_sf
+ More
IPR006068 ATPase_P-typ_cation-transptr_C
IPR023298 ATPase_P-typ_TM_dom_sf
IPR001757 P_typ_ATPase
IPR023299 ATPase_P-typ_cyto_dom_N
IPR030320 ATP2B1
IPR022141 ATP_Ca_trans_C
IPR036412 HAD-like_sf
IPR023214 HAD_sf
IPR006408 P-type_ATPase_IIB
IPR018303 ATPase_P-typ_P_site
IPR004014 ATPase_P-typ_cation-transptr_N
IPR034304 ATP2B4
IPR030325 ATP2B3
IPR006068 ATPase_P-typ_cation-transptr_C
IPR023298 ATPase_P-typ_TM_dom_sf
IPR001757 P_typ_ATPase
IPR023299 ATPase_P-typ_cyto_dom_N
IPR030320 ATP2B1
IPR022141 ATP_Ca_trans_C
IPR036412 HAD-like_sf
IPR023214 HAD_sf
IPR006408 P-type_ATPase_IIB
IPR018303 ATPase_P-typ_P_site
IPR004014 ATPase_P-typ_cation-transptr_N
IPR034304 ATP2B4
IPR030325 ATP2B3
Gene 3D
ProteinModelPortal
A0A232F311
V9I6Y1
A0A0K8W474
A0A0K8U0F8
A0A0K8UCZ8
A0A034W2K7
+ More
A0A034W3M2 A0A034W478 A0A0K8V849 A0A034W033 A0A1W4VHY3 A0A0N8NZM1 A0A0P8XIK1 A0A1W4VIP7 A0A1I8P3E0 A0A1A9WYB0 T1P9V5 A0A1I8M346 A0A1I8PR34 A0A1I8MG26 A0A1S4ERF4 A0A3Q0IU77 B4PW03 A0A0R1ECC4 A0A0Q5SDQ7 E6EK15 A0A0Q5SQA8 A0A1B0F9W3 A0A1I8PK10 A0A0L7R0N1 A0A0M8ZY79 A0A310S5N1
A0A034W3M2 A0A034W478 A0A0K8V849 A0A034W033 A0A1W4VHY3 A0A0N8NZM1 A0A0P8XIK1 A0A1W4VIP7 A0A1I8P3E0 A0A1A9WYB0 T1P9V5 A0A1I8M346 A0A1I8PR34 A0A1I8MG26 A0A1S4ERF4 A0A3Q0IU77 B4PW03 A0A0R1ECC4 A0A0Q5SDQ7 E6EK15 A0A0Q5SQA8 A0A1B0F9W3 A0A1I8PK10 A0A0L7R0N1 A0A0M8ZY79 A0A310S5N1
PDB
6A69
E-value=9.11273e-162,
Score=1466
Ontologies
GO
Topology
Subcellular location
Membrane
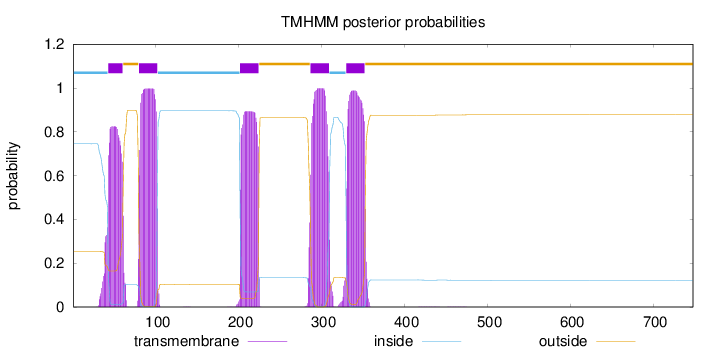
Length:
748
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
103.89829
Exp number, first 60 AAs:
16.03522
Total prob of N-in:
0.74528
POSSIBLE N-term signal
sequence
inside
1 - 42
TMhelix
43 - 60
outside
61 - 79
TMhelix
80 - 102
inside
103 - 201
TMhelix
202 - 224
outside
225 - 286
TMhelix
287 - 309
inside
310 - 329
TMhelix
330 - 352
outside
353 - 748
Population Genetic Test Statistics
Pi
234.508353
Theta
181.266469
Tajima's D
0.743714
CLR
0.351605
CSRT
0.590620468976551
Interpretation
Uncertain
