Gene
KWMTBOMO16122
Pre Gene Modal
BGIBMGA004513
Annotation
PREDICTED:_tyrosine-protein_phosphatase_69D_[Bombyx_mori]
Full name
Protein-tyrosine-phosphatase
+ More
Tyrosine-protein phosphatase 69D
Tyrosine-protein phosphatase 69D
Alternative Name
Protein-tyrosine-phosphate phosphohydrolase
Location in the cell
Nuclear Reliability : 2.81
Sequence
CDS
ATGTATCATAGACCTGACTTCAGTGGTACTTCCGCCATGACAATGGGTACTGTGGTCGGGAAGTGTTCCTTCGCGTTTTTAGTTTATTTGGCGTGTATTGAACTTTTGTATCCTGTTTTGGCTGAAGACAATGGAATTAGTTTAACAGCAAAAGGTGAAATCGGTAATGAATCAAATATTACTTGTGTTGTGCCCCTAGAAAATGCTACTGTTGAGTGGCAGTTTCAAGGAGAATCACTCAAAAAAGATAACAGACTACACATCACCGCATCTCAAGGTACACTGTCGAGGAAAGATCAAGATGGCTCATTGCAGAAGACTACAATTTCCACAATCATCATAACCAATACTACATCAGATTATGAAGGGAATTATACTTGTATAGCCAAGCAAGGAACCGAAGGACCCAGTATAAAGGAGTCTAGATTTCTGGACCTCAGTTTCCCAGGAAGGCTCATAAACACAACTAAAAGTCCTATCCGTGAAAATGCTACTAATCACAATAATGTCACTCTGGAATGCATATTTGAAGTTTATAAGCCTGGAACGGTCGTGTGGTCGAAGCGTGGCTCGGAAGACGACACCAGCAACCTGAACGATATAAAACAAGAGAATATAAACCTTAAACAAGTGAAGAGCTCCGTCGTGGTCCGCATCCGGAACGCGGACAACGACGACGGCTCGTACGTCTGCGAGGTGGACGACCACGTCACTAGCAGGCGGCTGTCCGGCGTCATCGACATCCACGTGTACGCGATGCCGGTCGTGGCCGTGGACAAGGCGGTCGCCGTCAACACCACCGAGGTCTTCCTCAACTGGACGGTCAAGTCCTACAGGCAGAAGATCGCGAACTACACCCTGATGTATCGCGTGAACTCGTCCAACGAAACGTTTCGGTTTTGCCGCGAACCGATAGACGTGAACAACACTTCGTTTGTGCTGAACGACCTGAACGCCTCCACGGAATACCAGCTGGTCCTGCAGGTCACCACCGCCTACGGCACGAGCAAGTCCAAAAACGTGATCGTGAGAACTTTGGACAAGGAACCGGTGTTTGTGCCGAACATTTCTATTAACGGTTTTTCCGCGACGTCAGTGACGATAGGCTGGCAACCCCCGCCCGAAGAAATATCGTCTCTGATCCACTACTACCTGTTAGAGGCGAGGAAGAAGGACGAGAACTCAACGAGGCGAGCCTACCACTCCGGAGGCATCATGAACCTGCCCTACATGTTCGACAACTTGGAGCCTCACAGCACTTACATATTTCGGGTGCGCGCGTGCTCGGAGATGACGCGGCGCTGCGGGCCCCGCTCGGCGGACATGCAGGCGGCCACGCTGGACGGCACGCCGGGCCGCCCCGCGCGTCTCCGCCTCCGCTGCGGCGCGCGCTCCCTGCGCGTGATGTGGGAGCCGCCCGAGCGACCCAACGCTGAGATCAAGGGCTACATGTTGGAATTGACAGGGAATGCTAATTACATAGATAAAACGGGCAAGGAGCAGAAGACCACCTGGGGGCCCTTGTCGAAGTTCACCTCCAACGATTCCAGGGAAGTCAGGTTCGAGGACCTGCAGCCCAACACGCAGTACACGGTGCGGCTGAGCGCCATGACGCGCACGCGGCGCCGCGGCGAGGAGGAGCTGCAGCGCTGCACCACGGCGCGGGCCGGCCCCGACCTGCCGCCCAGCCCGCGCTGGAGGAAGATATTAGACAACGATAATTATTATTTCAAAGTGTACATACCGCGTGTGTCGGAGAGAAACGGACCTATATGCTGCTACAGGGTGTACATGGTCCGCGTGGCGGGCGACGCGTCGCTGCGGGAACTGTCCCCGCTGCGCGCCGGACACTCCGCGCACCTCGACGCCTACGTCACCGACATCTTCTCCAACAAAAACTTCCCGCCGGATTCCGAGCTGGTGATGGGAGACGGGCGCTCGCACTACGACCCGGCGCGCGACCCCGCCCTCGCCGACGACAACTGCTCGCGCTGTCTCAAGGAACCCAAACGGTGA
Protein
MYHRPDFSGTSAMTMGTVVGKCSFAFLVYLACIELLYPVLAEDNGISLTAKGEIGNESNITCVVPLENATVEWQFQGESLKKDNRLHITASQGTLSRKDQDGSLQKTTISTIIITNTTSDYEGNYTCIAKQGTEGPSIKESRFLDLSFPGRLINTTKSPIRENATNHNNVTLECIFEVYKPGTVVWSKRGSEDDTSNLNDIKQENINLKQVKSSVVVRIRNADNDDGSYVCEVDDHVTSRRLSGVIDIHVYAMPVVAVDKAVAVNTTEVFLNWTVKSYRQKIANYTLMYRVNSSNETFRFCREPIDVNNTSFVLNDLNASTEYQLVLQVTTAYGTSKSKNVIVRTLDKEPVFVPNISINGFSATSVTIGWQPPPEEISSLIHYYLLEARKKDENSTRRAYHSGGIMNLPYMFDNLEPHSTYIFRVRACSEMTRRCGPRSADMQAATLDGTPGRPARLRLRCGARSLRVMWEPPERPNAEIKGYMLELTGNANYIDKTGKEQKTTWGPLSKFTSNDSREVRFEDLQPNTQYTVRLSAMTRTRRRGEEELQRCTTARAGPDLPPSPRWRKILDNDNYYFKVYIPRVSERNGPICCYRVYMVRVAGDASLRELSPLRAGHSAHLDAYVTDIFSNKNFPPDSELVMGDGRSHYDPARDPALADDNCSRCLKEPKR
Summary
Catalytic Activity
H2O + O-phospho-L-tyrosyl-[protein] = L-tyrosyl-[protein] + phosphate
Similarity
Belongs to the protein-tyrosine phosphatase family. Receptor class subfamily.
Keywords
Alternative splicing
Cell adhesion
Complete proteome
Disulfide bond
Glycoprotein
Hydrolase
Immunoglobulin domain
Membrane
Protein phosphatase
Receptor
Reference proteome
Repeat
Signal
Transmembrane
Transmembrane helix
Feature
chain Tyrosine-protein phosphatase 69D
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9J4S3
A0A212FIL0
A0A3S2NU76
A0A194RGQ7
A0A2H1WF51
A0A194QBE4
+ More
A0A2A4JGE0 K7IW60 A0A232F3N6 A0A067R8F5 A0A182YHJ0 A0A182LCE1 A0A336KPC3 A0A182XE51 A0A182HJ73 A0A182UQ71 B4L0P5 A0A1B0CK56 Q7QBG0 A0A182UBP6 A0A0L7QQQ1 A0A3B0JSF7 A0A182MFS8 A0A0J7KYF5 B0X101 A0A182RC55 A0A182PT15 B4LGG7 A0A182K464 A0A1Q3F8L9 Q2M0K8 A0A1Q3F8M0 C4IXZ1 A0A182QX88 A0A182JAQ7 W4VRM2 E2AHY3 W8BFL1 A0A1B6ISM3 A0A154P569 Q176N2 A0A1S4FD43 A0A182GNL7 A0A088A887 A0A182VWN8 B4HG53 A0A2J7Q3D3 A0A2J7Q3C3 A0A1B0BAB4 A0A1A9VMX8 A0A1B0G3J5 A0A1A9W4L0 A0A1B6KQ29 A0A0Q5UJ07 B3NCT8 A0A1A9YN25 A0A0J9RUH2 B4QRQ6 A0A1B6M0D4 A0A182N519 A0A1B0A2G4 A0A0M9A5X6 A0A034VDE1 B4IWZ0 P16620 P16620-2 A0A1W4XER9 A0A1W4X4E2 A0A0R1DXQ0 B4PGT9 A0A1W4XDU9 A0A1W4X4B1 A0A0T6AW03 A0A1W4UV77 A0A1W4UGK5 A0A1W4UG17 A0A310SHL4 A0A1I8MQR4 A0A084VAL3 A0A158NNE2 A0A1I8PYB7 A0A1I8PYC5 A0A151I388 A0A1Y1NLL3 A0A0L0BLG0 E2BAF9 A0A151J091 A0A151WUK2 A0A1Y1NGV2 A0A151JYX5 F4WXN1 A0A195CCJ6 B3M9P4 A0A2A3E8K9 A0A1Y1NGW3 E0VNQ0 A0A1J1IFK4 A0A2M4B973
A0A2A4JGE0 K7IW60 A0A232F3N6 A0A067R8F5 A0A182YHJ0 A0A182LCE1 A0A336KPC3 A0A182XE51 A0A182HJ73 A0A182UQ71 B4L0P5 A0A1B0CK56 Q7QBG0 A0A182UBP6 A0A0L7QQQ1 A0A3B0JSF7 A0A182MFS8 A0A0J7KYF5 B0X101 A0A182RC55 A0A182PT15 B4LGG7 A0A182K464 A0A1Q3F8L9 Q2M0K8 A0A1Q3F8M0 C4IXZ1 A0A182QX88 A0A182JAQ7 W4VRM2 E2AHY3 W8BFL1 A0A1B6ISM3 A0A154P569 Q176N2 A0A1S4FD43 A0A182GNL7 A0A088A887 A0A182VWN8 B4HG53 A0A2J7Q3D3 A0A2J7Q3C3 A0A1B0BAB4 A0A1A9VMX8 A0A1B0G3J5 A0A1A9W4L0 A0A1B6KQ29 A0A0Q5UJ07 B3NCT8 A0A1A9YN25 A0A0J9RUH2 B4QRQ6 A0A1B6M0D4 A0A182N519 A0A1B0A2G4 A0A0M9A5X6 A0A034VDE1 B4IWZ0 P16620 P16620-2 A0A1W4XER9 A0A1W4X4E2 A0A0R1DXQ0 B4PGT9 A0A1W4XDU9 A0A1W4X4B1 A0A0T6AW03 A0A1W4UV77 A0A1W4UGK5 A0A1W4UG17 A0A310SHL4 A0A1I8MQR4 A0A084VAL3 A0A158NNE2 A0A1I8PYB7 A0A1I8PYC5 A0A151I388 A0A1Y1NLL3 A0A0L0BLG0 E2BAF9 A0A151J091 A0A151WUK2 A0A1Y1NGV2 A0A151JYX5 F4WXN1 A0A195CCJ6 B3M9P4 A0A2A3E8K9 A0A1Y1NGW3 E0VNQ0 A0A1J1IFK4 A0A2M4B973
EC Number
3.1.3.48
Pubmed
EMBL
BABH01035520
AGBW02008361
OWR53575.1
RSAL01000078
RVE48717.1
KQ460207
+ More
KPJ16772.1 ODYU01008270 SOQ51708.1 KQ459249 KPJ02310.1 NWSH01001588 PCG70778.1 NNAY01001046 OXU25361.1 KK852812 KDR15856.1 UFQS01000322 UFQT01000322 SSX02860.1 SSX23227.1 APCN01002128 CH933809 EDW18122.2 AJWK01015759 AJWK01015760 AJWK01015761 AAAB01008879 EAA08408.5 KQ414786 KOC60952.1 OUUW01000009 SPP85055.1 AXCM01008699 LBMM01002057 KMQ95329.1 DS232250 EDS38393.1 CH940647 EDW70496.2 GFDL01011130 JAV23915.1 CH379069 EAL30922.1 GFDL01011131 JAV23914.1 BT083422 ACQ91626.1 AXCN02000177 GANO01001405 JAB58466.1 GL439602 EFN66973.1 GAMC01006450 JAC00106.1 GECU01017777 JAS89929.1 KQ434820 KZC07001.1 CH477385 EAT42115.1 JXUM01014645 JXUM01014646 JXUM01014647 JXUM01014648 JXUM01014649 JXUM01014650 KQ560409 KXJ82559.1 CH480815 EDW41299.1 NEVH01019068 PNF23077.1 PNF23076.1 JXJN01010887 JXJN01010888 CCAG010011629 GEBQ01026421 JAT13556.1 CH954178 KQS43863.1 EDV51594.1 CM002912 KMY99272.1 CM000363 EDX10270.1 KMY99271.1 GEBQ01010578 JAT29399.1 KQ435747 KOX76519.1 GAKP01019172 JAC39780.1 CH916366 EDV97391.1 M27699 AE014296 BT001531 CM000159 KRK01765.1 EDW94328.1 LJIG01022682 KRT79270.1 KQ759883 OAD62167.1 ATLV01004129 KE524190 KFB35007.1 ADTU01021299 ADTU01021300 ADTU01021301 ADTU01021302 ADTU01021303 ADTU01021304 KQ976489 KYM83471.1 GEZM01002835 JAV97136.1 JRES01001699 KNC20876.1 GL446721 EFN87321.1 KQ980644 KYN14909.1 KQ982730 KYQ51508.1 GEZM01002836 JAV97134.1 KQ981453 KYN41587.1 GL888426 EGI61031.1 KQ977935 KYM98579.1 CH902618 EDV39050.1 KZ288325 PBC28040.1 GEZM01002837 JAV97132.1 DS235341 EEB15006.1 CVRI01000047 CRK98538.1 GGFJ01000463 MBW49604.1
KPJ16772.1 ODYU01008270 SOQ51708.1 KQ459249 KPJ02310.1 NWSH01001588 PCG70778.1 NNAY01001046 OXU25361.1 KK852812 KDR15856.1 UFQS01000322 UFQT01000322 SSX02860.1 SSX23227.1 APCN01002128 CH933809 EDW18122.2 AJWK01015759 AJWK01015760 AJWK01015761 AAAB01008879 EAA08408.5 KQ414786 KOC60952.1 OUUW01000009 SPP85055.1 AXCM01008699 LBMM01002057 KMQ95329.1 DS232250 EDS38393.1 CH940647 EDW70496.2 GFDL01011130 JAV23915.1 CH379069 EAL30922.1 GFDL01011131 JAV23914.1 BT083422 ACQ91626.1 AXCN02000177 GANO01001405 JAB58466.1 GL439602 EFN66973.1 GAMC01006450 JAC00106.1 GECU01017777 JAS89929.1 KQ434820 KZC07001.1 CH477385 EAT42115.1 JXUM01014645 JXUM01014646 JXUM01014647 JXUM01014648 JXUM01014649 JXUM01014650 KQ560409 KXJ82559.1 CH480815 EDW41299.1 NEVH01019068 PNF23077.1 PNF23076.1 JXJN01010887 JXJN01010888 CCAG010011629 GEBQ01026421 JAT13556.1 CH954178 KQS43863.1 EDV51594.1 CM002912 KMY99272.1 CM000363 EDX10270.1 KMY99271.1 GEBQ01010578 JAT29399.1 KQ435747 KOX76519.1 GAKP01019172 JAC39780.1 CH916366 EDV97391.1 M27699 AE014296 BT001531 CM000159 KRK01765.1 EDW94328.1 LJIG01022682 KRT79270.1 KQ759883 OAD62167.1 ATLV01004129 KE524190 KFB35007.1 ADTU01021299 ADTU01021300 ADTU01021301 ADTU01021302 ADTU01021303 ADTU01021304 KQ976489 KYM83471.1 GEZM01002835 JAV97136.1 JRES01001699 KNC20876.1 GL446721 EFN87321.1 KQ980644 KYN14909.1 KQ982730 KYQ51508.1 GEZM01002836 JAV97134.1 KQ981453 KYN41587.1 GL888426 EGI61031.1 KQ977935 KYM98579.1 CH902618 EDV39050.1 KZ288325 PBC28040.1 GEZM01002837 JAV97132.1 DS235341 EEB15006.1 CVRI01000047 CRK98538.1 GGFJ01000463 MBW49604.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000053240
UP000053268
UP000218220
+ More
UP000002358 UP000215335 UP000027135 UP000076408 UP000075882 UP000076407 UP000075840 UP000075903 UP000009192 UP000092461 UP000007062 UP000075902 UP000053825 UP000268350 UP000075883 UP000036403 UP000002320 UP000075900 UP000075885 UP000008792 UP000075881 UP000001819 UP000075886 UP000075880 UP000000311 UP000076502 UP000008820 UP000069940 UP000249989 UP000005203 UP000075920 UP000001292 UP000235965 UP000092460 UP000078200 UP000092444 UP000091820 UP000008711 UP000092443 UP000000304 UP000075884 UP000092445 UP000053105 UP000001070 UP000000803 UP000192223 UP000002282 UP000192221 UP000095301 UP000030765 UP000005205 UP000095300 UP000078540 UP000037069 UP000008237 UP000078492 UP000075809 UP000078541 UP000007755 UP000078542 UP000007801 UP000242457 UP000009046 UP000183832
UP000002358 UP000215335 UP000027135 UP000076408 UP000075882 UP000076407 UP000075840 UP000075903 UP000009192 UP000092461 UP000007062 UP000075902 UP000053825 UP000268350 UP000075883 UP000036403 UP000002320 UP000075900 UP000075885 UP000008792 UP000075881 UP000001819 UP000075886 UP000075880 UP000000311 UP000076502 UP000008820 UP000069940 UP000249989 UP000005203 UP000075920 UP000001292 UP000235965 UP000092460 UP000078200 UP000092444 UP000091820 UP000008711 UP000092443 UP000000304 UP000075884 UP000092445 UP000053105 UP000001070 UP000000803 UP000192223 UP000002282 UP000192221 UP000095301 UP000030765 UP000005205 UP000095300 UP000078540 UP000037069 UP000008237 UP000078492 UP000075809 UP000078541 UP000007755 UP000078542 UP000007801 UP000242457 UP000009046 UP000183832
Interpro
IPR036179
Ig-like_dom_sf
+ More
IPR036116 FN3_sf
IPR013151 Immunoglobulin
IPR013783 Ig-like_fold
IPR003961 FN3_dom
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013098 Ig_I-set
IPR000242 PTPase_domain
IPR000387 TYR_PHOSPHATASE_dom
IPR016130 Tyr_Pase_AS
IPR029021 Prot-tyrosine_phosphatase-like
IPR003595 Tyr_Pase_cat
IPR003598 Ig_sub2
IPR003329 Cytidylyl_trans
IPR029044 Nucleotide-diphossugar_trans
IPR036116 FN3_sf
IPR013151 Immunoglobulin
IPR013783 Ig-like_fold
IPR003961 FN3_dom
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013098 Ig_I-set
IPR000242 PTPase_domain
IPR000387 TYR_PHOSPHATASE_dom
IPR016130 Tyr_Pase_AS
IPR029021 Prot-tyrosine_phosphatase-like
IPR003595 Tyr_Pase_cat
IPR003598 Ig_sub2
IPR003329 Cytidylyl_trans
IPR029044 Nucleotide-diphossugar_trans
Gene 3D
CDD
ProteinModelPortal
H9J4S3
A0A212FIL0
A0A3S2NU76
A0A194RGQ7
A0A2H1WF51
A0A194QBE4
+ More
A0A2A4JGE0 K7IW60 A0A232F3N6 A0A067R8F5 A0A182YHJ0 A0A182LCE1 A0A336KPC3 A0A182XE51 A0A182HJ73 A0A182UQ71 B4L0P5 A0A1B0CK56 Q7QBG0 A0A182UBP6 A0A0L7QQQ1 A0A3B0JSF7 A0A182MFS8 A0A0J7KYF5 B0X101 A0A182RC55 A0A182PT15 B4LGG7 A0A182K464 A0A1Q3F8L9 Q2M0K8 A0A1Q3F8M0 C4IXZ1 A0A182QX88 A0A182JAQ7 W4VRM2 E2AHY3 W8BFL1 A0A1B6ISM3 A0A154P569 Q176N2 A0A1S4FD43 A0A182GNL7 A0A088A887 A0A182VWN8 B4HG53 A0A2J7Q3D3 A0A2J7Q3C3 A0A1B0BAB4 A0A1A9VMX8 A0A1B0G3J5 A0A1A9W4L0 A0A1B6KQ29 A0A0Q5UJ07 B3NCT8 A0A1A9YN25 A0A0J9RUH2 B4QRQ6 A0A1B6M0D4 A0A182N519 A0A1B0A2G4 A0A0M9A5X6 A0A034VDE1 B4IWZ0 P16620 P16620-2 A0A1W4XER9 A0A1W4X4E2 A0A0R1DXQ0 B4PGT9 A0A1W4XDU9 A0A1W4X4B1 A0A0T6AW03 A0A1W4UV77 A0A1W4UGK5 A0A1W4UG17 A0A310SHL4 A0A1I8MQR4 A0A084VAL3 A0A158NNE2 A0A1I8PYB7 A0A1I8PYC5 A0A151I388 A0A1Y1NLL3 A0A0L0BLG0 E2BAF9 A0A151J091 A0A151WUK2 A0A1Y1NGV2 A0A151JYX5 F4WXN1 A0A195CCJ6 B3M9P4 A0A2A3E8K9 A0A1Y1NGW3 E0VNQ0 A0A1J1IFK4 A0A2M4B973
A0A2A4JGE0 K7IW60 A0A232F3N6 A0A067R8F5 A0A182YHJ0 A0A182LCE1 A0A336KPC3 A0A182XE51 A0A182HJ73 A0A182UQ71 B4L0P5 A0A1B0CK56 Q7QBG0 A0A182UBP6 A0A0L7QQQ1 A0A3B0JSF7 A0A182MFS8 A0A0J7KYF5 B0X101 A0A182RC55 A0A182PT15 B4LGG7 A0A182K464 A0A1Q3F8L9 Q2M0K8 A0A1Q3F8M0 C4IXZ1 A0A182QX88 A0A182JAQ7 W4VRM2 E2AHY3 W8BFL1 A0A1B6ISM3 A0A154P569 Q176N2 A0A1S4FD43 A0A182GNL7 A0A088A887 A0A182VWN8 B4HG53 A0A2J7Q3D3 A0A2J7Q3C3 A0A1B0BAB4 A0A1A9VMX8 A0A1B0G3J5 A0A1A9W4L0 A0A1B6KQ29 A0A0Q5UJ07 B3NCT8 A0A1A9YN25 A0A0J9RUH2 B4QRQ6 A0A1B6M0D4 A0A182N519 A0A1B0A2G4 A0A0M9A5X6 A0A034VDE1 B4IWZ0 P16620 P16620-2 A0A1W4XER9 A0A1W4X4E2 A0A0R1DXQ0 B4PGT9 A0A1W4XDU9 A0A1W4X4B1 A0A0T6AW03 A0A1W4UV77 A0A1W4UGK5 A0A1W4UG17 A0A310SHL4 A0A1I8MQR4 A0A084VAL3 A0A158NNE2 A0A1I8PYB7 A0A1I8PYC5 A0A151I388 A0A1Y1NLL3 A0A0L0BLG0 E2BAF9 A0A151J091 A0A151WUK2 A0A1Y1NGV2 A0A151JYX5 F4WXN1 A0A195CCJ6 B3M9P4 A0A2A3E8K9 A0A1Y1NGW3 E0VNQ0 A0A1J1IFK4 A0A2M4B973
PDB
4PBX
E-value=3.45231e-14,
Score=193
Ontologies
GO
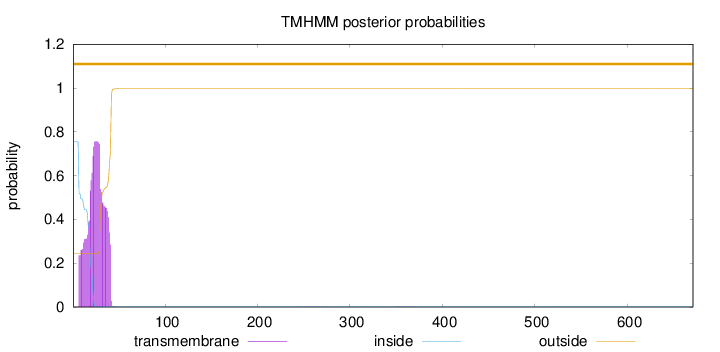
Topology
Subcellular location
Membrane
Length:
671
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
16.61885
Exp number, first 60 AAs:
16.60647
Total prob of N-in:
0.75419
POSSIBLE N-term signal
sequence
outside
1 - 671
Population Genetic Test Statistics
Pi
219.35547
Theta
182.103278
Tajima's D
0.66046
CLR
0.386198
CSRT
0.560321983900805
Interpretation
Uncertain
