Gene
KWMTBOMO16115
Pre Gene Modal
BGIBMGA004538
Annotation
PREDICTED:_sugar_transporter_ERD6-like_6_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.783
Sequence
CDS
ATGTCTGGTACTGGACGATGGACCACGCCGTTCTTGAAACAGTGCTTTGCGGTATCGAGTGTGGCTCTCAACGTAATGTCATACGGTATGGTTTTCGGATTTAACGCATCACTGTTCTCAGACTTGAGGAAGAGTCGAGAGATACCCTTGGACAGGGATTCAGAGTCATGGTTGGCATCACTGATCGGAATAACTACACTAATAGGATGTTTCTCCACCTCAATAATCATAGACACAATTGGCAGAAGACCTGCACTTCTCATCAGCTCAACCCTCATGGTCAGCGGTTGGATAGGATTCAGTTTGGCCTCATCATTTCCTGCCTTACTATTAGCAAAGATATTTCAAGGAATGTCTATCGGTCTTGGGACCACAATGGGCGGTATTTTAATTGCAGAATACAGCAGTCCCAAATACCGTGGATCTCTTATAGCAACAATACCTGCTGCGTTCCTGGTCGTGTTTTATTATATGGTACCGGAAACTAAAGATAAGACTCTTCAAGAAATAGAAGATAAACTCAGAGGATATTCTATAGCTCGTCGTGATGAAACTTTGAAATTAAAAGACGCTGAACTATGA
Protein
MSGTGRWTTPFLKQCFAVSSVALNVMSYGMVFGFNASLFSDLRKSREIPLDRDSESWLASLIGITTLIGCFSTSIIIDTIGRRPALLISSTLMVSGWIGFSLASSFPALLLAKIFQGMSIGLGTTMGGILIAEYSSPKYRGSLIATIPAAFLVVFYYMVPETKDKTLQEIEDKLRGYSIARRDETLKLKDAEL
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9J4R7
H9J4S0
A0A3S2P2D1
A0A2A4KAY2
A0A2W1BG84
A0A0N1IPS2
+ More
H9J4X7 A0A3S2NWF9 A0A2H1WVP2 H9J4X1 A0A2A4JTG8 A0A023PNK9 A0A212F161 A0A2H1W936 A0A194QJ23 A0A0N0PDT5 A0A2A4JT91 H9J4S1 A0A0N1IG19 A0A2H1WFL6 A0A2A4JUU3 A0A2A4JNG3 A0A2H1WAU6 A0A2W1BD78 A0A194QJI7 A0A0N1IPZ0 A0A194QJ40 A0A212FPF6 A0A212F9B1 A0A194QJ30 H9J533 A0A194QJJ6 A0A2A4JSU4 A0A194QKX0 A0A194QHU0 A0A3S2LUZ6 A0A0N1IIX6 A0A3S2LEF2 A0A1L8DF47 A0A1L8DFA5 A0A2A4J748 H9J4X3 A0A182WIV2 A0A0N1IDG3 A0A194QJ80 A0A2W1BCE3 A0A2W1BF54 H9J539 A0A1B0CWH0 A0A0A8J8L7 A0A2W1BFI0 A0A2A4JVB9 A0A336MJD0 A0A336M7L2 K7J6E5 A0A232ESL4 A0A2H1WB04
H9J4X7 A0A3S2NWF9 A0A2H1WVP2 H9J4X1 A0A2A4JTG8 A0A023PNK9 A0A212F161 A0A2H1W936 A0A194QJ23 A0A0N0PDT5 A0A2A4JT91 H9J4S1 A0A0N1IG19 A0A2H1WFL6 A0A2A4JUU3 A0A2A4JNG3 A0A2H1WAU6 A0A2W1BD78 A0A194QJI7 A0A0N1IPZ0 A0A194QJ40 A0A212FPF6 A0A212F9B1 A0A194QJ30 H9J533 A0A194QJJ6 A0A2A4JSU4 A0A194QKX0 A0A194QHU0 A0A3S2LUZ6 A0A0N1IIX6 A0A3S2LEF2 A0A1L8DF47 A0A1L8DFA5 A0A2A4J748 H9J4X3 A0A182WIV2 A0A0N1IDG3 A0A194QJ80 A0A2W1BCE3 A0A2W1BF54 H9J539 A0A1B0CWH0 A0A0A8J8L7 A0A2W1BFI0 A0A2A4JVB9 A0A336MJD0 A0A336M7L2 K7J6E5 A0A232ESL4 A0A2H1WB04
EMBL
BABH01035544
BABH01035538
BABH01035539
RSAL01000045
RVE50592.1
NWSH01000008
+ More
PCG80953.1 KZ150249 PZC71836.1 KQ460077 KPJ17881.1 BABH01025717 RVE50589.1 ODYU01011356 SOQ57052.1 BABH01025731 NWSH01000704 PCG74682.1 KJ439227 KZ150491 AHX25878.1 PZC70688.1 AGBW02010956 OWR47467.1 ODYU01007122 SOQ49615.1 KQ458671 KPJ05558.1 KPJ17885.1 PCG74683.1 BABH01035527 BABH01035528 BABH01035529 KPJ17891.1 ODYU01008354 SOQ51875.1 NWSH01000578 PCG75536.1 NWSH01001042 PCG72950.1 SOQ49614.1 PZC71835.1 KPJ05554.1 KQ460126 KPJ17589.1 KPJ05573.1 AGBW02002687 OWR55626.1 AGBW02009614 OWR50324.1 KPJ05563.1 BABH01038850 KPJ05559.1 PCG74684.1 KPJ05555.1 KQ458981 KPJ04505.1 RSAL01000181 RVE44925.1 KPJ17884.1 RVE44926.1 GFDF01009026 JAV05058.1 GFDF01009027 JAV05057.1 NWSH01002771 PCG67556.1 BABH01025723 KQ459158 KPJ03590.1 KPJ05572.1 PZC70686.1 PZC71837.1 BABH01038877 BABH01038878 BABH01038879 BABH01038880 BABH01038881 AJWK01032461 AB901078 BAQ02378.1 KZ150133 PZC73021.1 NWSH01000610 PCG75342.1 UFQT01001316 SSX30005.1 UFQT01000224 SSX22028.1 NNAY01002414 OXU21332.1 ODYU01007347 SOQ50032.1
PCG80953.1 KZ150249 PZC71836.1 KQ460077 KPJ17881.1 BABH01025717 RVE50589.1 ODYU01011356 SOQ57052.1 BABH01025731 NWSH01000704 PCG74682.1 KJ439227 KZ150491 AHX25878.1 PZC70688.1 AGBW02010956 OWR47467.1 ODYU01007122 SOQ49615.1 KQ458671 KPJ05558.1 KPJ17885.1 PCG74683.1 BABH01035527 BABH01035528 BABH01035529 KPJ17891.1 ODYU01008354 SOQ51875.1 NWSH01000578 PCG75536.1 NWSH01001042 PCG72950.1 SOQ49614.1 PZC71835.1 KPJ05554.1 KQ460126 KPJ17589.1 KPJ05573.1 AGBW02002687 OWR55626.1 AGBW02009614 OWR50324.1 KPJ05563.1 BABH01038850 KPJ05559.1 PCG74684.1 KPJ05555.1 KQ458981 KPJ04505.1 RSAL01000181 RVE44925.1 KPJ17884.1 RVE44926.1 GFDF01009026 JAV05058.1 GFDF01009027 JAV05057.1 NWSH01002771 PCG67556.1 BABH01025723 KQ459158 KPJ03590.1 KPJ05572.1 PZC70686.1 PZC71837.1 BABH01038877 BABH01038878 BABH01038879 BABH01038880 BABH01038881 AJWK01032461 AB901078 BAQ02378.1 KZ150133 PZC73021.1 NWSH01000610 PCG75342.1 UFQT01001316 SSX30005.1 UFQT01000224 SSX22028.1 NNAY01002414 OXU21332.1 ODYU01007347 SOQ50032.1
Proteomes
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9J4R7
H9J4S0
A0A3S2P2D1
A0A2A4KAY2
A0A2W1BG84
A0A0N1IPS2
+ More
H9J4X7 A0A3S2NWF9 A0A2H1WVP2 H9J4X1 A0A2A4JTG8 A0A023PNK9 A0A212F161 A0A2H1W936 A0A194QJ23 A0A0N0PDT5 A0A2A4JT91 H9J4S1 A0A0N1IG19 A0A2H1WFL6 A0A2A4JUU3 A0A2A4JNG3 A0A2H1WAU6 A0A2W1BD78 A0A194QJI7 A0A0N1IPZ0 A0A194QJ40 A0A212FPF6 A0A212F9B1 A0A194QJ30 H9J533 A0A194QJJ6 A0A2A4JSU4 A0A194QKX0 A0A194QHU0 A0A3S2LUZ6 A0A0N1IIX6 A0A3S2LEF2 A0A1L8DF47 A0A1L8DFA5 A0A2A4J748 H9J4X3 A0A182WIV2 A0A0N1IDG3 A0A194QJ80 A0A2W1BCE3 A0A2W1BF54 H9J539 A0A1B0CWH0 A0A0A8J8L7 A0A2W1BFI0 A0A2A4JVB9 A0A336MJD0 A0A336M7L2 K7J6E5 A0A232ESL4 A0A2H1WB04
H9J4X7 A0A3S2NWF9 A0A2H1WVP2 H9J4X1 A0A2A4JTG8 A0A023PNK9 A0A212F161 A0A2H1W936 A0A194QJ23 A0A0N0PDT5 A0A2A4JT91 H9J4S1 A0A0N1IG19 A0A2H1WFL6 A0A2A4JUU3 A0A2A4JNG3 A0A2H1WAU6 A0A2W1BD78 A0A194QJI7 A0A0N1IPZ0 A0A194QJ40 A0A212FPF6 A0A212F9B1 A0A194QJ30 H9J533 A0A194QJJ6 A0A2A4JSU4 A0A194QKX0 A0A194QHU0 A0A3S2LUZ6 A0A0N1IIX6 A0A3S2LEF2 A0A1L8DF47 A0A1L8DFA5 A0A2A4J748 H9J4X3 A0A182WIV2 A0A0N1IDG3 A0A194QJ80 A0A2W1BCE3 A0A2W1BF54 H9J539 A0A1B0CWH0 A0A0A8J8L7 A0A2W1BFI0 A0A2A4JVB9 A0A336MJD0 A0A336M7L2 K7J6E5 A0A232ESL4 A0A2H1WB04
Ontologies
GO
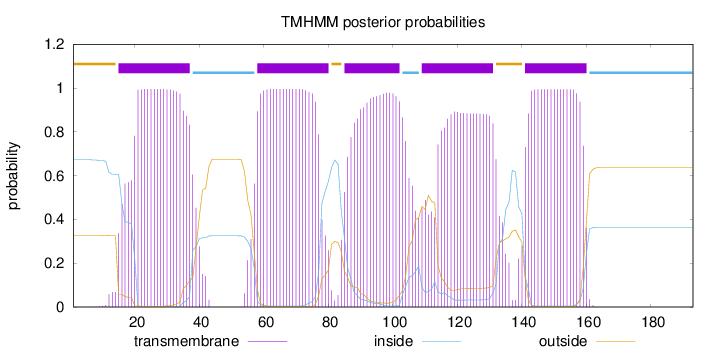
Topology
Length:
193
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
102.23035
Exp number, first 60 AAs:
25.66699
Total prob of N-in:
0.67316
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 37
inside
38 - 57
TMhelix
58 - 80
outside
81 - 84
TMhelix
85 - 102
inside
103 - 108
TMhelix
109 - 131
outside
132 - 140
TMhelix
141 - 160
inside
161 - 193
Population Genetic Test Statistics
Pi
216.869053
Theta
139.679711
Tajima's D
1.303097
CLR
0.147815
CSRT
0.735713214339283
Interpretation
Uncertain
