Gene
KWMTBOMO16114
Annotation
PREDICTED:_facilitated_trehalose_transporter_Tret1-like_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.928
Sequence
CDS
ATGCCTCGTCGCAGACAATGGATGACGCCATTTATAAAACAGTGTTTCGTCACATCAGGTGTGACACTCAATATGTTGACACACGGCTTCATGTACGGATTCAATACGGGACTGTTCGCACAGTTGAGGAAGACTAAAGAAATACAGTTGGATTTGGAGTTAGAGTCTTGGCTGGCGTCGTCAACCAGCTTATCATTCATCGTTGGATCGCTATTCAGTTCAATATTCATGGATCGGTTCGGCAGGAGACCTGCATTCATAATTGCATCGGCCACCATGATAATCAGCTGGTTAATATTCATTGCAGCTAAATCCTTCACCGCTTTATTAGTTGCCAAAATATTTCAAGGCTTCTCTGCAGCGGCTGGATCTGTTTCATTACCAAATATTGTGGCCGGAGAAATATTCCCTTTGGCAAATAGAAGCGCTTGTGGAATGATTTGTAACATAACATTTTCATTATACATGTTCGTTAATATCAAAAACGTTCCTTATATCTTTTCTGGGGCTGGGGTCAGCGGTGTTTTTTTTATGAACGCCGCCCTGCTTTCATATGCCCTCGGCATGACAATGTACACGTTACCGGAAACAAAAGATAAAACGCTCTTAGAAATAGAACACATATTAAGAGGATGTCCTATTGCTGATGATGATGAGACAATGATTAATCTGAAAGAACATAATGAGACAAATTTCTAA
Protein
MPRRRQWMTPFIKQCFVTSGVTLNMLTHGFMYGFNTGLFAQLRKTKEIQLDLELESWLASSTSLSFIVGSLFSSIFMDRFGRRPAFIIASATMIISWLIFIAAKSFTALLVAKIFQGFSAAAGSVSLPNIVAGEIFPLANRSACGMICNITFSLYMFVNIKNVPYIFSGAGVSGVFFMNAALLSYALGMTMYTLPETKDKTLLEIEHILRGCPIADDDETMINLKEHNETNF
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9J4R9
H9J4S0
A0A2A4JT91
H9J4X7
A0A2A4JTG8
A0A023PNK9
+ More
A0A2H1W936 A0A2W1BBH7 A0A2W1BD78 A0A2A4JUU3 A0A0N1IPS2 A0A2A4JKJ9 H9J017 A0A0N1IPZ0 A0A0N1IDG3 A0A2H1WKZ9 A0A194QKX0 A0A0N0PEE0 A0A0N0PEP7 A0A1S4FT49 A0A0N0PAH4 A0A2A4JJR5 A0A2H1W9B8 A0A0D6MUP8 J9KWZ6 A0A2S3W2F7 A0A2V4R238 A0A2H8TN79 D5QII2 A0A143WL99 A0A318Q7D5 A0A0L7LRT2 A0A336MDU0 A0A336LS95 B4KG70
A0A2H1W936 A0A2W1BBH7 A0A2W1BD78 A0A2A4JUU3 A0A0N1IPS2 A0A2A4JKJ9 H9J017 A0A0N1IPZ0 A0A0N1IDG3 A0A2H1WKZ9 A0A194QKX0 A0A0N0PEE0 A0A0N0PEP7 A0A1S4FT49 A0A0N0PAH4 A0A2A4JJR5 A0A2H1W9B8 A0A0D6MUP8 J9KWZ6 A0A2S3W2F7 A0A2V4R238 A0A2H8TN79 D5QII2 A0A143WL99 A0A318Q7D5 A0A0L7LRT2 A0A336MDU0 A0A336LS95 B4KG70
EMBL
BABH01035541
BABH01035538
BABH01035539
NWSH01000704
PCG74683.1
BABH01025717
+ More
PCG74682.1 KJ439227 KZ150491 AHX25878.1 PZC70688.1 ODYU01007122 SOQ49615.1 PZC70687.1 KZ150249 PZC71835.1 NWSH01000578 PCG75536.1 KQ460077 KPJ17881.1 NWSH01001160 PCG72306.1 BABH01018801 KQ460126 KPJ17589.1 KQ459158 KPJ03590.1 ODYU01009373 SOQ53753.1 KQ458671 KPJ05555.1 KQ459890 KPJ19576.1 KPJ19577.1 KQ458842 KPJ04897.1 PCG72307.1 SOQ49613.1 BAMU01000017 GAN57452.1 ABLF02025479 ABLF02025480 POTC01000012 POF63050.1 NKUB01000002 PYD70956.1 GFXV01003635 MBW15440.1 ADTV01000060 EFG83150.1 FBVP01000001 CUW48336.1 NKUD01000004 PYD73328.1 JTDY01000230 KOB78170.1 UFQT01001016 SSX28504.1 UFQS01000083 UFQT01000083 SSW99067.1 SSX19449.1 CH933807 EDW11057.1
PCG74682.1 KJ439227 KZ150491 AHX25878.1 PZC70688.1 ODYU01007122 SOQ49615.1 PZC70687.1 KZ150249 PZC71835.1 NWSH01000578 PCG75536.1 KQ460077 KPJ17881.1 NWSH01001160 PCG72306.1 BABH01018801 KQ460126 KPJ17589.1 KQ459158 KPJ03590.1 ODYU01009373 SOQ53753.1 KQ458671 KPJ05555.1 KQ459890 KPJ19576.1 KPJ19577.1 KQ458842 KPJ04897.1 PCG72307.1 SOQ49613.1 BAMU01000017 GAN57452.1 ABLF02025479 ABLF02025480 POTC01000012 POF63050.1 NKUB01000002 PYD70956.1 GFXV01003635 MBW15440.1 ADTV01000060 EFG83150.1 FBVP01000001 CUW48336.1 NKUD01000004 PYD73328.1 JTDY01000230 KOB78170.1 UFQT01001016 SSX28504.1 UFQS01000083 UFQT01000083 SSW99067.1 SSX19449.1 CH933807 EDW11057.1
Proteomes
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9J4R9
H9J4S0
A0A2A4JT91
H9J4X7
A0A2A4JTG8
A0A023PNK9
+ More
A0A2H1W936 A0A2W1BBH7 A0A2W1BD78 A0A2A4JUU3 A0A0N1IPS2 A0A2A4JKJ9 H9J017 A0A0N1IPZ0 A0A0N1IDG3 A0A2H1WKZ9 A0A194QKX0 A0A0N0PEE0 A0A0N0PEP7 A0A1S4FT49 A0A0N0PAH4 A0A2A4JJR5 A0A2H1W9B8 A0A0D6MUP8 J9KWZ6 A0A2S3W2F7 A0A2V4R238 A0A2H8TN79 D5QII2 A0A143WL99 A0A318Q7D5 A0A0L7LRT2 A0A336MDU0 A0A336LS95 B4KG70
A0A2H1W936 A0A2W1BBH7 A0A2W1BD78 A0A2A4JUU3 A0A0N1IPS2 A0A2A4JKJ9 H9J017 A0A0N1IPZ0 A0A0N1IDG3 A0A2H1WKZ9 A0A194QKX0 A0A0N0PEE0 A0A0N0PEP7 A0A1S4FT49 A0A0N0PAH4 A0A2A4JJR5 A0A2H1W9B8 A0A0D6MUP8 J9KWZ6 A0A2S3W2F7 A0A2V4R238 A0A2H8TN79 D5QII2 A0A143WL99 A0A318Q7D5 A0A0L7LRT2 A0A336MDU0 A0A336LS95 B4KG70
PDB
4LDS
E-value=0.000608201,
Score=99
Ontologies
GO
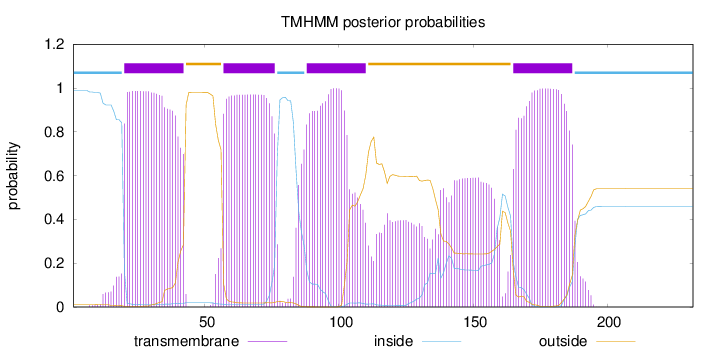
Topology
Length:
232
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
107.45045
Exp number, first 60 AAs:
26.57953
Total prob of N-in:
0.98911
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 42
outside
43 - 56
TMhelix
57 - 76
inside
77 - 87
TMhelix
88 - 110
outside
111 - 164
TMhelix
165 - 187
inside
188 - 232
Population Genetic Test Statistics
Pi
178.683179
Theta
183.223457
Tajima's D
-0.34069
CLR
344.67027
CSRT
0.27863606819659
Interpretation
Uncertain
