Gene
KWMTBOMO16110
Pre Gene Modal
BGIBMGA004622
Annotation
PREDICTED:_cathepsin_L-like_proteinase_isoform_X1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.548
Sequence
CDS
ATGTCTGAGATACTTTTCAGACGAACGTTGATATGGAAAAAAAAATTAGAAAGTTCCAGAAAACCAAAAATTTCTGCAGCAATTCACAATAAATTGTATTCATCTACGCACCACGAGATGGCAGCGCTAATTAAATGGCGGCAGAACTTGAGACGGGTTGCGAGACACAACAGAGAGTACTTAGCGGGAATACAGTCGTACTCATTACATTTGAATCATTTTGGAGATATGCATGTCACAGAATACTTCGGCAAAGTTTTGAAGCTGATCAAAGCATTCCCGCTCTTCGATCCAGCTGAAGACCACCACAAGACCGCGTATAGGCATAACAGACGCTGCAAAGTGCCGAAAAGGATAGATTGGCGGGATCAAGGTTTCAAGCCGAGACGAGAGGAGCAATGGCAATGTGGTGCTTGTTATGCCTTTGCCGTAACCCATGCTTTGCAGGCTCAGCTATACAAGAGACACGGGGAGTGGAATGAGCTCAGTCCTCAACAAATCGTGGACTGCAGCATTAAAGACGGCAATATGGGATGCGACGGAGGATCCTTGAGGGGAGCCCTACGTTATGCAGCCAGAGAGGGCCTGGTTATGGAGTCCCATTATCCGTATGTTGGAAAGAAAGGATACTGCAGATACGATAGCAATTTGGTGCGCGCCAGACCGCGGCGTTGGGCCACACTTCCGTCAGGCGATGAAGAAGCAATGGAGAAGGCCTTAGCTACTGTAGGTCCTCTAGCTGTCGCCGTTAATGCTGCCCCATTCACTTTTCAGCTTTACAGAAGCGGCGTTTACGATGACCCCTTTTGCGTGTCGTGGCATCTGAATCATGCTATGTTATTAGTCGGGTATACTCAGGATTATTGGATACTGCTCAACTGGTGGGGCAGAAATTGGGGTGAAGATGGATACATGAGAATACGAAGAGGCTTAAACCGATGTGGCGTCGCCAACATGGCAACCTATGTAGAATTGTAA
Protein
MSEILFRRTLIWKKKLESSRKPKISAAIHNKLYSSTHHEMAALIKWRQNLRRVARHNREYLAGIQSYSLHLNHFGDMHVTEYFGKVLKLIKAFPLFDPAEDHHKTAYRHNRRCKVPKRIDWRDQGFKPRREEQWQCGACYAFAVTHALQAQLYKRHGEWNELSPQQIVDCSIKDGNMGCDGGSLRGALRYAAREGLVMESHYPYVGKKGYCRYDSNLVRARPRRWATLPSGDEEAMEKALATVGPLAVAVNAAPFTFQLYRSGVYDDPFCVSWHLNHAMLLVGYTQDYWILLNWWGRNWGEDGYMRIRRGLNRCGVANMATYVEL
Summary
Similarity
Belongs to the peptidase C1 family.
Uniprot
A0FDR3
H9J532
A0A2A4J7N8
A0A212ESU6
A0A3S2LKM4
J7HLS2
+ More
A0A0N1IIE6 A0A0N1ICC4 A0A2H1VZI0 A0A146MDS5 W8C7M2 A0A0C9QUA6 B4L7D4 B4G780 K7IXW4 A0A034WW51 Q9VKY4 B4Q973 A0A0J9R0X8 E2C3B6 B4HWL8 B3N9M3 A0A3B0KDD6 B4NZA8 A0A1Y1JRC9 A0A026W1K8 D6W755 B4JZP3 A0A0L7QLG5 A0A3L8D729 B3MPS2 A0A1B6D4B7 T1HW30 A0A154PL20 A0A0Q5WM32 A0A158P2P4 B4MEY5 A0A195BCX3 A0A151IGC4 A0A195DCI0 D0Z790 A0A0R1DL52 A0A088AUV9 A0A195F3W3 A0A0Q9X488 A0A0L0BUD7 E2AI46 A0A1W4USH4 N6T961 A0A336KK13 U4TWX9 E9J7R5 W8CEC3 A0A1S4ECC8 A0A1B6H716 A0A2S2N8N2 A0A1J1I6X8 A0A1B0DQ40 A0A3B0K6M8 A0A2R2YUN1 E4W4G9 W5K309 U6JMH6 A2BF64 Q4QRH8 Q566T8 F2E5Q3 G9F9V4 A7RRL9 A0A3S3QVU1 A0A3L7IFH5 H9GEA5 H0VE49 A0A267H1I6 A0A3Q1BJS2 Q90324 A0A1I8GVB7 H3BEF4 A0A146VNF7 A0A1U8CIN5 H6QXS8 A0T069 A0A3B1KAS0 H8PGF3 A0A3B4AD44 A0A2I4BAA8 A0A3B4C7Z3 A0A3Q1CNG8 A0A3M7PN10 A0A098M1C2 A0A3P9MK65 A0A1I7ZU59 A0A3P8SQI4 A0A3P8TP03 A0A1V4KMP4
A0A0N1IIE6 A0A0N1ICC4 A0A2H1VZI0 A0A146MDS5 W8C7M2 A0A0C9QUA6 B4L7D4 B4G780 K7IXW4 A0A034WW51 Q9VKY4 B4Q973 A0A0J9R0X8 E2C3B6 B4HWL8 B3N9M3 A0A3B0KDD6 B4NZA8 A0A1Y1JRC9 A0A026W1K8 D6W755 B4JZP3 A0A0L7QLG5 A0A3L8D729 B3MPS2 A0A1B6D4B7 T1HW30 A0A154PL20 A0A0Q5WM32 A0A158P2P4 B4MEY5 A0A195BCX3 A0A151IGC4 A0A195DCI0 D0Z790 A0A0R1DL52 A0A088AUV9 A0A195F3W3 A0A0Q9X488 A0A0L0BUD7 E2AI46 A0A1W4USH4 N6T961 A0A336KK13 U4TWX9 E9J7R5 W8CEC3 A0A1S4ECC8 A0A1B6H716 A0A2S2N8N2 A0A1J1I6X8 A0A1B0DQ40 A0A3B0K6M8 A0A2R2YUN1 E4W4G9 W5K309 U6JMH6 A2BF64 Q4QRH8 Q566T8 F2E5Q3 G9F9V4 A7RRL9 A0A3S3QVU1 A0A3L7IFH5 H9GEA5 H0VE49 A0A267H1I6 A0A3Q1BJS2 Q90324 A0A1I8GVB7 H3BEF4 A0A146VNF7 A0A1U8CIN5 H6QXS8 A0T069 A0A3B1KAS0 H8PGF3 A0A3B4AD44 A0A2I4BAA8 A0A3B4C7Z3 A0A3Q1CNG8 A0A3M7PN10 A0A098M1C2 A0A3P9MK65 A0A1I7ZU59 A0A3P8SQI4 A0A3P8TP03 A0A1V4KMP4
Pubmed
19121390
22118469
26354079
26823975
24495485
17994087
+ More
20075255 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 20798317 17550304 28004739 24508170 18362917 19820115 30249741 21347285 26108605 23537049 21282665 25329095 25875018 23594743 21415278 17615350 29704459 21881562 21993624 26392545 9215903 26319212 25463417 30375419 17554307 26392177
20075255 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 20798317 17550304 28004739 24508170 18362917 19820115 30249741 21347285 26108605 23537049 21282665 25329095 25875018 23594743 21415278 17615350 29704459 21881562 21993624 26392545 9215903 26319212 25463417 30375419 17554307 26392177
EMBL
DQ985605
ABJ97193.1
BABH01038834
NWSH01002787
PCG67534.1
AGBW02012704
+ More
OWR44576.1 RSAL01000078 RVE48727.1 JN400437 AFQ01140.1 KQ459890 KPJ19578.1 KQ458842 KPJ04898.1 ODYU01005395 SOQ46187.1 GDHC01000681 JAQ17948.1 GAMC01000447 JAC06109.1 GBYB01007324 JAG77091.1 CH933813 EDW10928.1 CH479180 EDW28334.1 GAKP01000919 JAC58033.1 AE014134 AAF52922.2 CM000361 EDX04515.1 CM002910 KMY89499.1 GL452318 EFN77562.1 CH480818 EDW52413.1 CH954177 EDV58518.2 OUUW01000006 SPP81668.1 CM000157 EDW88803.2 GEZM01102813 JAV51779.1 KK107488 EZA49955.1 KQ971307 EFA11596.1 CH916380 EDV90909.1 KQ414934 KOC59356.1 QOIP01000012 RLU15678.1 CH902620 EDV32320.1 GEDC01016771 JAS20527.1 ACPB03001684 ACPB03001685 KQ434954 KZC12523.1 KQS70431.1 ADTU01001283 ADTU01001284 CH940665 EDW71086.2 KQ976522 KYM82062.1 KQ977726 KYN00271.1 KQ980989 KYN10542.1 CM000831 ACY70535.1 KRJ98039.1 KQ981855 KYN34774.1 CH964284 KRG00324.1 JRES01001323 KNC23636.1 GL439639 EFN66897.1 APGK01047235 KB741077 ENN74258.1 UFQS01000526 UFQT01000526 SSX04621.1 SSX24984.1 KB631792 ERL86114.1 GL768602 EFZ11138.1 GAMC01000446 JAC06110.1 GECU01037276 JAS70430.1 GGMR01000886 MBY13505.1 CVRI01000038 CRK94177.1 AJVK01018842 AJVK01018843 AJVK01018844 AJVK01018845 SPP81669.1 KM522788 AKN80414.1 EU643472 ACF48469.1 HG710159 CDJ26735.1 BX248517 BC096862 AAH96862.1 BC093339 BC164362 AAH93339.1 AAI64362.1 AK371477 BAK02675.1 JN099751 AEM76722.1 DS469532 EDO45852.1 NCKU01000620 RWS14794.1 RAZU01000058 RLQ76893.1 AAWZ02033459 AAKN02038469 NIVC01000061 PAA92133.1 L30111 AAA49207.1 AFYH01037257 GCES01067339 JAR18984.1 FN667958 CBJ56264.1 EF067847 ABK63481.1 FN687459 CBK52289.1 REGN01009714 RNA00502.1 GBSI01000475 JAC96021.1 LSYS01002801 OPJ85685.1
OWR44576.1 RSAL01000078 RVE48727.1 JN400437 AFQ01140.1 KQ459890 KPJ19578.1 KQ458842 KPJ04898.1 ODYU01005395 SOQ46187.1 GDHC01000681 JAQ17948.1 GAMC01000447 JAC06109.1 GBYB01007324 JAG77091.1 CH933813 EDW10928.1 CH479180 EDW28334.1 GAKP01000919 JAC58033.1 AE014134 AAF52922.2 CM000361 EDX04515.1 CM002910 KMY89499.1 GL452318 EFN77562.1 CH480818 EDW52413.1 CH954177 EDV58518.2 OUUW01000006 SPP81668.1 CM000157 EDW88803.2 GEZM01102813 JAV51779.1 KK107488 EZA49955.1 KQ971307 EFA11596.1 CH916380 EDV90909.1 KQ414934 KOC59356.1 QOIP01000012 RLU15678.1 CH902620 EDV32320.1 GEDC01016771 JAS20527.1 ACPB03001684 ACPB03001685 KQ434954 KZC12523.1 KQS70431.1 ADTU01001283 ADTU01001284 CH940665 EDW71086.2 KQ976522 KYM82062.1 KQ977726 KYN00271.1 KQ980989 KYN10542.1 CM000831 ACY70535.1 KRJ98039.1 KQ981855 KYN34774.1 CH964284 KRG00324.1 JRES01001323 KNC23636.1 GL439639 EFN66897.1 APGK01047235 KB741077 ENN74258.1 UFQS01000526 UFQT01000526 SSX04621.1 SSX24984.1 KB631792 ERL86114.1 GL768602 EFZ11138.1 GAMC01000446 JAC06110.1 GECU01037276 JAS70430.1 GGMR01000886 MBY13505.1 CVRI01000038 CRK94177.1 AJVK01018842 AJVK01018843 AJVK01018844 AJVK01018845 SPP81669.1 KM522788 AKN80414.1 EU643472 ACF48469.1 HG710159 CDJ26735.1 BX248517 BC096862 AAH96862.1 BC093339 BC164362 AAH93339.1 AAI64362.1 AK371477 BAK02675.1 JN099751 AEM76722.1 DS469532 EDO45852.1 NCKU01000620 RWS14794.1 RAZU01000058 RLQ76893.1 AAWZ02033459 AAKN02038469 NIVC01000061 PAA92133.1 L30111 AAA49207.1 AFYH01037257 GCES01067339 JAR18984.1 FN667958 CBJ56264.1 EF067847 ABK63481.1 FN687459 CBK52289.1 REGN01009714 RNA00502.1 GBSI01000475 JAC96021.1 LSYS01002801 OPJ85685.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053240
UP000053268
+ More
UP000009192 UP000008744 UP000002358 UP000000803 UP000000304 UP000008237 UP000001292 UP000008711 UP000268350 UP000002282 UP000053097 UP000007266 UP000001070 UP000053825 UP000279307 UP000007801 UP000015103 UP000076502 UP000005205 UP000008792 UP000078540 UP000078542 UP000078492 UP000005203 UP000078541 UP000007798 UP000037069 UP000000311 UP000192221 UP000019118 UP000030742 UP000079169 UP000183832 UP000092462 UP000018467 UP000000437 UP000001593 UP000285301 UP000273346 UP000001646 UP000005447 UP000215902 UP000257160 UP000095280 UP000008672 UP000189706 UP000261520 UP000192220 UP000261440 UP000276133 UP000265180 UP000095287 UP000265080 UP000190648
UP000009192 UP000008744 UP000002358 UP000000803 UP000000304 UP000008237 UP000001292 UP000008711 UP000268350 UP000002282 UP000053097 UP000007266 UP000001070 UP000053825 UP000279307 UP000007801 UP000015103 UP000076502 UP000005205 UP000008792 UP000078540 UP000078542 UP000078492 UP000005203 UP000078541 UP000007798 UP000037069 UP000000311 UP000192221 UP000019118 UP000030742 UP000079169 UP000183832 UP000092462 UP000018467 UP000000437 UP000001593 UP000285301 UP000273346 UP000001646 UP000005447 UP000215902 UP000257160 UP000095280 UP000008672 UP000189706 UP000261520 UP000192220 UP000261440 UP000276133 UP000265180 UP000095287 UP000265080 UP000190648
Interpro
ProteinModelPortal
A0FDR3
H9J532
A0A2A4J7N8
A0A212ESU6
A0A3S2LKM4
J7HLS2
+ More
A0A0N1IIE6 A0A0N1ICC4 A0A2H1VZI0 A0A146MDS5 W8C7M2 A0A0C9QUA6 B4L7D4 B4G780 K7IXW4 A0A034WW51 Q9VKY4 B4Q973 A0A0J9R0X8 E2C3B6 B4HWL8 B3N9M3 A0A3B0KDD6 B4NZA8 A0A1Y1JRC9 A0A026W1K8 D6W755 B4JZP3 A0A0L7QLG5 A0A3L8D729 B3MPS2 A0A1B6D4B7 T1HW30 A0A154PL20 A0A0Q5WM32 A0A158P2P4 B4MEY5 A0A195BCX3 A0A151IGC4 A0A195DCI0 D0Z790 A0A0R1DL52 A0A088AUV9 A0A195F3W3 A0A0Q9X488 A0A0L0BUD7 E2AI46 A0A1W4USH4 N6T961 A0A336KK13 U4TWX9 E9J7R5 W8CEC3 A0A1S4ECC8 A0A1B6H716 A0A2S2N8N2 A0A1J1I6X8 A0A1B0DQ40 A0A3B0K6M8 A0A2R2YUN1 E4W4G9 W5K309 U6JMH6 A2BF64 Q4QRH8 Q566T8 F2E5Q3 G9F9V4 A7RRL9 A0A3S3QVU1 A0A3L7IFH5 H9GEA5 H0VE49 A0A267H1I6 A0A3Q1BJS2 Q90324 A0A1I8GVB7 H3BEF4 A0A146VNF7 A0A1U8CIN5 H6QXS8 A0T069 A0A3B1KAS0 H8PGF3 A0A3B4AD44 A0A2I4BAA8 A0A3B4C7Z3 A0A3Q1CNG8 A0A3M7PN10 A0A098M1C2 A0A3P9MK65 A0A1I7ZU59 A0A3P8SQI4 A0A3P8TP03 A0A1V4KMP4
A0A0N1IIE6 A0A0N1ICC4 A0A2H1VZI0 A0A146MDS5 W8C7M2 A0A0C9QUA6 B4L7D4 B4G780 K7IXW4 A0A034WW51 Q9VKY4 B4Q973 A0A0J9R0X8 E2C3B6 B4HWL8 B3N9M3 A0A3B0KDD6 B4NZA8 A0A1Y1JRC9 A0A026W1K8 D6W755 B4JZP3 A0A0L7QLG5 A0A3L8D729 B3MPS2 A0A1B6D4B7 T1HW30 A0A154PL20 A0A0Q5WM32 A0A158P2P4 B4MEY5 A0A195BCX3 A0A151IGC4 A0A195DCI0 D0Z790 A0A0R1DL52 A0A088AUV9 A0A195F3W3 A0A0Q9X488 A0A0L0BUD7 E2AI46 A0A1W4USH4 N6T961 A0A336KK13 U4TWX9 E9J7R5 W8CEC3 A0A1S4ECC8 A0A1B6H716 A0A2S2N8N2 A0A1J1I6X8 A0A1B0DQ40 A0A3B0K6M8 A0A2R2YUN1 E4W4G9 W5K309 U6JMH6 A2BF64 Q4QRH8 Q566T8 F2E5Q3 G9F9V4 A7RRL9 A0A3S3QVU1 A0A3L7IFH5 H9GEA5 H0VE49 A0A267H1I6 A0A3Q1BJS2 Q90324 A0A1I8GVB7 H3BEF4 A0A146VNF7 A0A1U8CIN5 H6QXS8 A0T069 A0A3B1KAS0 H8PGF3 A0A3B4AD44 A0A2I4BAA8 A0A3B4C7Z3 A0A3Q1CNG8 A0A3M7PN10 A0A098M1C2 A0A3P9MK65 A0A1I7ZU59 A0A3P8SQI4 A0A3P8TP03 A0A1V4KMP4
PDB
7PCK
E-value=3.01892e-52,
Score=518
Ontologies
GO
GO:0008234
GO:0022857
GO:0016021
GO:0004197
GO:0005615
GO:0005764
GO:0051603
GO:0016485
GO:0005518
GO:0097067
GO:0043236
GO:0002250
GO:0001968
GO:0030574
GO:0043394
GO:0034769
GO:0010447
GO:0048002
GO:0005770
GO:2001259
GO:0006508
GO:0016020
GO:0022891
GO:0001678
GO:0005536
GO:0046835
GO:0006281
GO:0006259
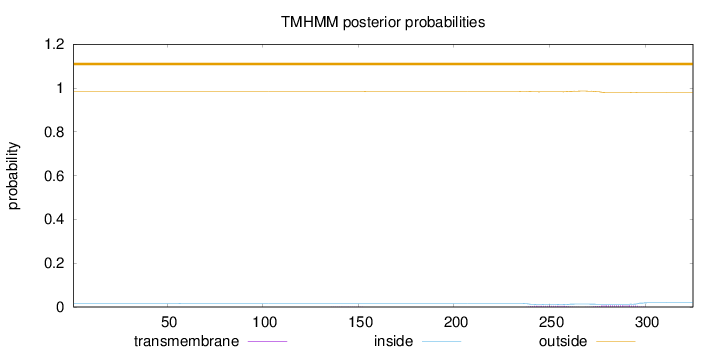
Topology
Length:
325
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.29018
Exp number, first 60 AAs:
0.0001
Total prob of N-in:
0.01542
outside
1 - 325
Population Genetic Test Statistics
Pi
33.833139
Theta
25.967532
Tajima's D
2.092986
CLR
0.399207
CSRT
0.900604969751512
Interpretation
Uncertain
