Gene
KWMTBOMO16102
Pre Gene Modal
BGIBMGA004623
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_facilitated_trehalose_transporter_Tret1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.982
Sequence
CDS
ATGGAGTCTTTTATAAAACAGGCCTTCGTCGTGAGCGGCGCTGCACTGAACATAGCAGGACACGGCTGTGCTCATGGCTACCCAGCTGTGCTCTTTGCGCAAATCGAAAGCAGCAAAGGCGACTTCAAACTCAGCGACCACGACACGTCATGGATTGCTTCAGTTGTCGGCGTTATGGGAATAGTGGGTAACTTCATATCGCCGGTTCTAATGACGAAGTATGGCCGTCAAAAAGCTCATCTCCTCACAACTATACCAGCTCTACTCGGCTGGCTGATATTCGTATTCGGAAACTCTGTCCCTCTATTCATTCTGGCGAGGCTACTCCACGGTCTGGCTTTAGGGCTAAGAACACCTTTAGCAGCGATATTAGTGGCAGAGTATACGGACCCAAAGTACAGGGGAGCGTTCTTGGGGACATTCGCCATATCACTAGGTCTCGGGATCTTCTTGTCACATCTTTGGGGAGCGCATTTAACGTGGAAAATGACTGCTGTGGTTTGTTCCATGTTCCCGTTGATCGCCATGGCGATAATATCTTTTTCTCCGGAGTCGCCGAGCTGGCTGGTCTCCAAAGGTAGGTACGAAGAGGCTAGGAAGTCATTTAGGTGGATTCGAGGCGGAGGTCCCGTACAAATGGAGGAGTTCGAAGCGATGGTTAATGCGCAGAAGGAAGAAGTGGCATCGGAAAAAAACGAACCGATACCAAAAAGGAATGGAATCTTCAAAACCGCGATAAATTCGATAAAGAGCGTCCTCAGAGTATTCAAGAAGAAAGAGTTTTATAAGCCGGCCGTCATAGCGATCAGCATGTTGGTCGTTTTCGAGTTCGGAGGAGCCCATATGTTCGCAGCCTACGGCAACACGATCCTCCAAGCCGTTTTGGACAAAGAAAACGCCACAGACGTGGCGTGGCAACTGACCGTGATGGACTTACTCCGCACGATCTGCGCGTTCTTGGCCATCTTCCTTTTGAAATACTGCAAAAGAAGAACGATATTATTCACCAGTGGCCTCACAACGGTTCTATCGCTCGCTTTTATATCTAGTTTTATTTACATGAGAAAAGCCGGTCTCTTCACGGCGACTTGGCTGCTGGAGCTAATGCCGATGTCCCTGATGATATTCTACACGGTCTCGTTTTGTCTTGGCCTCGTTCCATTGAACTGGGTTATATGTGGCGAAGTATTCCCGTTAGCTTACCGAAGTCTAGGATCTACTTTGTCAACGTCATTTTTGACGCCGTCTTTTGTGATCTCCATGAAGACGGCTCCACATTTGTATTCTTCTGTGGGGGTCGAGGGAGCGTTCCTGCTGTACTCGGCCATTTTAACGTTCTTCTTGGTTATAATGTATATTTTACTACCCGAAACCAAAGACAGGACCCTTCAAGATATCGAAGAAAGTTTTAAAGGCAAAAAGAACGCAGACCCGGAAGTACAAATGACGCTTATAAATAAAACCGATAGTGTGTTAGTCACGTAA
Protein
MESFIKQAFVVSGAALNIAGHGCAHGYPAVLFAQIESSKGDFKLSDHDTSWIASVVGVMGIVGNFISPVLMTKYGRQKAHLLTTIPALLGWLIFVFGNSVPLFILARLLHGLALGLRTPLAAILVAEYTDPKYRGAFLGTFAISLGLGIFLSHLWGAHLTWKMTAVVCSMFPLIAMAIISFSPESPSWLVSKGRYEEARKSFRWIRGGGPVQMEEFEAMVNAQKEEVASEKNEPIPKRNGIFKTAINSIKSVLRVFKKKEFYKPAVIAISMLVVFEFGGAHMFAAYGNTILQAVLDKENATDVAWQLTVMDLLRTICAFLAIFLLKYCKRRTILFTSGLTTVLSLAFISSFIYMRKAGLFTATWLLELMPMSLMIFYTVSFCLGLVPLNWVICGEVFPLAYRSLGSTLSTSFLTPSFVISMKTAPHLYSSVGVEGAFLLYSAILTFFLVIMYILLPETKDRTLQDIEESFKGKKNADPEVQMTLINKTDSVLVT
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9J533
A0A2A4JNG3
A0A0N1IG19
A0A194QJ30
A0A2H1X0D6
A0A0L7LRT2
+ More
A0A212FPF6 H9J525 A0A2A4JTG8 A0A023PNK9 A0A2A4JT91 A0A2H1W936 A0A212F9B1 A0A194QJ23 A0A2H1WFL6 H9J4X7 A0A2W1BD78 A0A3S2LUZ6 A0A2A4JUU3 A0A2H1WAU6 A0A194QHU0 A0A194QJI7 A0A0N1IPS2 A0A212F161 A0A3S2P2D1 A0A2A4J748 A0A0L7KZR3 A0A2H1WVP2 A0A194QJ40 A0A2W1BG84 A0A2A4KAY2 A0A2H1WB04 A0A2W1BFI0 A0A212FJ74 A0A2A4JSU4 A0A0N1I9K7 A0A194QPY2 A0A0N1IHN6 A0A212FCC0 A0A2A4KA42 H9J4X0 H9J4X2 E0X906 A0A194QKX0 A0A2W1B9E8 A0A2H1WK46 A0A182KD28 H9J4R6 A0A2H1WFM1 A0A182RRX2 A0A182WKH8 A0A212F9C9 A0A3S2NWF9 A0A182XN09 A0A182VI49 A0A182NHP5 A0A2W1BDG5 A0A182MLS9 A0A0L7L8D8 A0A182LJ94 A0A182Y162 A0A182PHI4 A0A2A4JU31 A0A194QJ63 A0A1S4GQI2 H9J4S1 A0A194QJ80 A0A182IUY7 A0A3S2LEF2 Q7Q6Y6 A0A194QKY0 B0W5I2 A0A182U3G7 H9J4X1 A0A2A4KB29 A0A2A4JU29 A0A084VC86 A0A182HWW7 A0A139WGJ0 A0A0L7L7N1 A0A3S2NLE7 A0A2H1V0I3 A0A182GTB3 A0A0L7L8F3 A0A182GCQ0 A0A0N1IPZ8 A0A2W1BF54 A0A194R9K8 A0A336MJD0 Q17LS5 A0A336M7L2 W5JEJ5 A0A2H1W9B8 H9J4X3 A0A182FDK9 A0A1J1IVY8 A0A212FCB5 A0A194QEH9 B0W5I1 A0A2H1WKZ9
A0A212FPF6 H9J525 A0A2A4JTG8 A0A023PNK9 A0A2A4JT91 A0A2H1W936 A0A212F9B1 A0A194QJ23 A0A2H1WFL6 H9J4X7 A0A2W1BD78 A0A3S2LUZ6 A0A2A4JUU3 A0A2H1WAU6 A0A194QHU0 A0A194QJI7 A0A0N1IPS2 A0A212F161 A0A3S2P2D1 A0A2A4J748 A0A0L7KZR3 A0A2H1WVP2 A0A194QJ40 A0A2W1BG84 A0A2A4KAY2 A0A2H1WB04 A0A2W1BFI0 A0A212FJ74 A0A2A4JSU4 A0A0N1I9K7 A0A194QPY2 A0A0N1IHN6 A0A212FCC0 A0A2A4KA42 H9J4X0 H9J4X2 E0X906 A0A194QKX0 A0A2W1B9E8 A0A2H1WK46 A0A182KD28 H9J4R6 A0A2H1WFM1 A0A182RRX2 A0A182WKH8 A0A212F9C9 A0A3S2NWF9 A0A182XN09 A0A182VI49 A0A182NHP5 A0A2W1BDG5 A0A182MLS9 A0A0L7L8D8 A0A182LJ94 A0A182Y162 A0A182PHI4 A0A2A4JU31 A0A194QJ63 A0A1S4GQI2 H9J4S1 A0A194QJ80 A0A182IUY7 A0A3S2LEF2 Q7Q6Y6 A0A194QKY0 B0W5I2 A0A182U3G7 H9J4X1 A0A2A4KB29 A0A2A4JU29 A0A084VC86 A0A182HWW7 A0A139WGJ0 A0A0L7L7N1 A0A3S2NLE7 A0A2H1V0I3 A0A182GTB3 A0A0L7L8F3 A0A182GCQ0 A0A0N1IPZ8 A0A2W1BF54 A0A194R9K8 A0A336MJD0 Q17LS5 A0A336M7L2 W5JEJ5 A0A2H1W9B8 H9J4X3 A0A182FDK9 A0A1J1IVY8 A0A212FCB5 A0A194QEH9 B0W5I1 A0A2H1WKZ9
Pubmed
EMBL
BABH01038850
NWSH01001042
PCG72950.1
KQ460077
KPJ17891.1
KQ458671
+ More
KPJ05563.1 ODYU01012470 SOQ58795.1 JTDY01000230 KOB78170.1 AGBW02002687 OWR55626.1 BABH01038881 NWSH01000704 PCG74682.1 KJ439227 KZ150491 AHX25878.1 PZC70688.1 PCG74683.1 ODYU01007122 SOQ49615.1 AGBW02009614 OWR50324.1 KPJ05558.1 ODYU01008354 SOQ51875.1 BABH01025717 KZ150249 PZC71835.1 RSAL01000181 RVE44925.1 NWSH01000578 PCG75536.1 SOQ49614.1 KQ458981 KPJ04505.1 KPJ05554.1 KPJ17881.1 AGBW02010956 OWR47467.1 RSAL01000045 RVE50592.1 NWSH01002771 PCG67556.1 JTDY01004123 KOB68566.1 ODYU01011356 SOQ57052.1 KPJ05573.1 PZC71836.1 NWSH01000008 PCG80953.1 ODYU01007347 SOQ50032.1 KZ150133 PZC73021.1 AGBW02008317 OWR53759.1 PCG74684.1 KPJ17880.1 KPJ05561.1 KPJ17887.1 AGBW02009205 OWR51384.1 PCG80949.1 BABH01025733 BABH01025724 GQ871756 ADM86147.1 KPJ05555.1 PZC71838.1 ODYU01009098 SOQ53272.1 BABH01035548 SOQ51878.1 OWR50323.1 RVE50589.1 PZC71834.1 AXCM01002831 JTDY01002352 KOB71610.1 PCG75535.1 KPJ05557.1 AAAB01008960 BABH01035527 BABH01035528 BABH01035529 KPJ05572.1 RVE44926.1 EAA11457.4 KPJ05560.1 DS231843 EDS35465.1 BABH01025731 PCG80950.1 PCG75537.1 ATLV01010625 KE524589 KFB35580.1 APCN01001553 KQ971345 KYB27009.1 JTDY01002470 KOB71349.1 RVE50591.1 ODYU01000118 SOQ34347.1 JXUM01086634 KQ563580 KXJ73638.1 KOB71609.1 JXUM01009472 KQ560284 KXJ83288.1 KPJ17883.1 PZC71837.1 KQ460761 KPJ12541.1 UFQT01001316 SSX30005.1 CH477213 EAT47626.1 UFQT01000224 SSX22028.1 ADMH02001735 ETN61249.1 SOQ49613.1 BABH01025723 CVRI01000059 CRL03302.1 OWR51386.1 KQ459299 KPJ01871.1 EDS35464.1 ODYU01009373 SOQ53753.1
KPJ05563.1 ODYU01012470 SOQ58795.1 JTDY01000230 KOB78170.1 AGBW02002687 OWR55626.1 BABH01038881 NWSH01000704 PCG74682.1 KJ439227 KZ150491 AHX25878.1 PZC70688.1 PCG74683.1 ODYU01007122 SOQ49615.1 AGBW02009614 OWR50324.1 KPJ05558.1 ODYU01008354 SOQ51875.1 BABH01025717 KZ150249 PZC71835.1 RSAL01000181 RVE44925.1 NWSH01000578 PCG75536.1 SOQ49614.1 KQ458981 KPJ04505.1 KPJ05554.1 KPJ17881.1 AGBW02010956 OWR47467.1 RSAL01000045 RVE50592.1 NWSH01002771 PCG67556.1 JTDY01004123 KOB68566.1 ODYU01011356 SOQ57052.1 KPJ05573.1 PZC71836.1 NWSH01000008 PCG80953.1 ODYU01007347 SOQ50032.1 KZ150133 PZC73021.1 AGBW02008317 OWR53759.1 PCG74684.1 KPJ17880.1 KPJ05561.1 KPJ17887.1 AGBW02009205 OWR51384.1 PCG80949.1 BABH01025733 BABH01025724 GQ871756 ADM86147.1 KPJ05555.1 PZC71838.1 ODYU01009098 SOQ53272.1 BABH01035548 SOQ51878.1 OWR50323.1 RVE50589.1 PZC71834.1 AXCM01002831 JTDY01002352 KOB71610.1 PCG75535.1 KPJ05557.1 AAAB01008960 BABH01035527 BABH01035528 BABH01035529 KPJ05572.1 RVE44926.1 EAA11457.4 KPJ05560.1 DS231843 EDS35465.1 BABH01025731 PCG80950.1 PCG75537.1 ATLV01010625 KE524589 KFB35580.1 APCN01001553 KQ971345 KYB27009.1 JTDY01002470 KOB71349.1 RVE50591.1 ODYU01000118 SOQ34347.1 JXUM01086634 KQ563580 KXJ73638.1 KOB71609.1 JXUM01009472 KQ560284 KXJ83288.1 KPJ17883.1 PZC71837.1 KQ460761 KPJ12541.1 UFQT01001316 SSX30005.1 CH477213 EAT47626.1 UFQT01000224 SSX22028.1 ADMH02001735 ETN61249.1 SOQ49613.1 BABH01025723 CVRI01000059 CRL03302.1 OWR51386.1 KQ459299 KPJ01871.1 EDS35464.1 ODYU01009373 SOQ53753.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000037510
UP000007151
+ More
UP000283053 UP000075881 UP000075900 UP000075920 UP000076407 UP000075903 UP000075884 UP000075883 UP000075882 UP000076408 UP000075885 UP000075880 UP000007062 UP000002320 UP000075902 UP000030765 UP000075840 UP000007266 UP000069940 UP000249989 UP000008820 UP000000673 UP000069272 UP000183832
UP000283053 UP000075881 UP000075900 UP000075920 UP000076407 UP000075903 UP000075884 UP000075883 UP000075882 UP000076408 UP000075885 UP000075880 UP000007062 UP000002320 UP000075902 UP000030765 UP000075840 UP000007266 UP000069940 UP000249989 UP000008820 UP000000673 UP000069272 UP000183832
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9J533
A0A2A4JNG3
A0A0N1IG19
A0A194QJ30
A0A2H1X0D6
A0A0L7LRT2
+ More
A0A212FPF6 H9J525 A0A2A4JTG8 A0A023PNK9 A0A2A4JT91 A0A2H1W936 A0A212F9B1 A0A194QJ23 A0A2H1WFL6 H9J4X7 A0A2W1BD78 A0A3S2LUZ6 A0A2A4JUU3 A0A2H1WAU6 A0A194QHU0 A0A194QJI7 A0A0N1IPS2 A0A212F161 A0A3S2P2D1 A0A2A4J748 A0A0L7KZR3 A0A2H1WVP2 A0A194QJ40 A0A2W1BG84 A0A2A4KAY2 A0A2H1WB04 A0A2W1BFI0 A0A212FJ74 A0A2A4JSU4 A0A0N1I9K7 A0A194QPY2 A0A0N1IHN6 A0A212FCC0 A0A2A4KA42 H9J4X0 H9J4X2 E0X906 A0A194QKX0 A0A2W1B9E8 A0A2H1WK46 A0A182KD28 H9J4R6 A0A2H1WFM1 A0A182RRX2 A0A182WKH8 A0A212F9C9 A0A3S2NWF9 A0A182XN09 A0A182VI49 A0A182NHP5 A0A2W1BDG5 A0A182MLS9 A0A0L7L8D8 A0A182LJ94 A0A182Y162 A0A182PHI4 A0A2A4JU31 A0A194QJ63 A0A1S4GQI2 H9J4S1 A0A194QJ80 A0A182IUY7 A0A3S2LEF2 Q7Q6Y6 A0A194QKY0 B0W5I2 A0A182U3G7 H9J4X1 A0A2A4KB29 A0A2A4JU29 A0A084VC86 A0A182HWW7 A0A139WGJ0 A0A0L7L7N1 A0A3S2NLE7 A0A2H1V0I3 A0A182GTB3 A0A0L7L8F3 A0A182GCQ0 A0A0N1IPZ8 A0A2W1BF54 A0A194R9K8 A0A336MJD0 Q17LS5 A0A336M7L2 W5JEJ5 A0A2H1W9B8 H9J4X3 A0A182FDK9 A0A1J1IVY8 A0A212FCB5 A0A194QEH9 B0W5I1 A0A2H1WKZ9
A0A212FPF6 H9J525 A0A2A4JTG8 A0A023PNK9 A0A2A4JT91 A0A2H1W936 A0A212F9B1 A0A194QJ23 A0A2H1WFL6 H9J4X7 A0A2W1BD78 A0A3S2LUZ6 A0A2A4JUU3 A0A2H1WAU6 A0A194QHU0 A0A194QJI7 A0A0N1IPS2 A0A212F161 A0A3S2P2D1 A0A2A4J748 A0A0L7KZR3 A0A2H1WVP2 A0A194QJ40 A0A2W1BG84 A0A2A4KAY2 A0A2H1WB04 A0A2W1BFI0 A0A212FJ74 A0A2A4JSU4 A0A0N1I9K7 A0A194QPY2 A0A0N1IHN6 A0A212FCC0 A0A2A4KA42 H9J4X0 H9J4X2 E0X906 A0A194QKX0 A0A2W1B9E8 A0A2H1WK46 A0A182KD28 H9J4R6 A0A2H1WFM1 A0A182RRX2 A0A182WKH8 A0A212F9C9 A0A3S2NWF9 A0A182XN09 A0A182VI49 A0A182NHP5 A0A2W1BDG5 A0A182MLS9 A0A0L7L8D8 A0A182LJ94 A0A182Y162 A0A182PHI4 A0A2A4JU31 A0A194QJ63 A0A1S4GQI2 H9J4S1 A0A194QJ80 A0A182IUY7 A0A3S2LEF2 Q7Q6Y6 A0A194QKY0 B0W5I2 A0A182U3G7 H9J4X1 A0A2A4KB29 A0A2A4JU29 A0A084VC86 A0A182HWW7 A0A139WGJ0 A0A0L7L7N1 A0A3S2NLE7 A0A2H1V0I3 A0A182GTB3 A0A0L7L8F3 A0A182GCQ0 A0A0N1IPZ8 A0A2W1BF54 A0A194R9K8 A0A336MJD0 Q17LS5 A0A336M7L2 W5JEJ5 A0A2H1W9B8 H9J4X3 A0A182FDK9 A0A1J1IVY8 A0A212FCB5 A0A194QEH9 B0W5I1 A0A2H1WKZ9
PDB
6H7D
E-value=7.6603e-17,
Score=214
Ontologies
GO
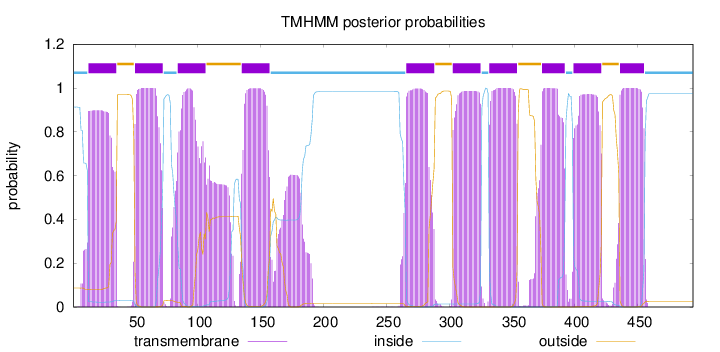
Topology
Length:
494
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
241.91177
Exp number, first 60 AAs:
31.56052
Total prob of N-in:
0.91326
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 49
TMhelix
50 - 72
inside
73 - 83
TMhelix
84 - 106
outside
107 - 134
TMhelix
135 - 157
inside
158 - 265
TMhelix
266 - 288
outside
289 - 302
TMhelix
303 - 325
inside
326 - 331
TMhelix
332 - 354
outside
355 - 373
TMhelix
374 - 392
inside
393 - 398
TMhelix
399 - 421
outside
422 - 435
TMhelix
436 - 455
inside
456 - 494
Population Genetic Test Statistics
Pi
234.772127
Theta
198.844269
Tajima's D
0.652952
CLR
0.169565
CSRT
0.56087195640218
Interpretation
Uncertain
