Gene
KWMTBOMO16101
Pre Gene Modal
BGIBMGA004619
Annotation
PREDICTED:_glycosyltransferase_isoform_X1_[Bombyx_mori]
Full name
Hexosyltransferase
Location in the cell
PlasmaMembrane Reliability : 1.935
Sequence
CDS
ATGACGGAATCTCAGCGGCCGAAAGAAAGAATTGAAATGATCACTCTACATAAGATACTATTTGTTTTTTGCTTTTTGTCTTTATCGCTAAACTATTATATATCGACAAAACCTATGCTAGACATTGAAAACCCAACTTTTTCTCTGATTCAACCTAATGAAACGATCACTCCGAAATTTAGTGATAATTACGACCAGTTGATTGATATAAAGAAATTCTCATTCAAAATCAATCCTAAACCATGTAAACGTTACCCAGAAGGTTTTCTACTTATGATAATAGTTTCCTCCAACCCCTTGAATTATGAAAACAGGCTTGTCATCAGAAAAACGTGGGGACAGACAGATGAGGCCACGAACATTGTATTTTTGGTTGGAGAGACAGATAATGTTACTGTATCACAGAAAATACAGGAAGAAAGTGTGACATATGGTGATATAGTGCAAGGTAACTTTAAAGATGCATACCATAACATGACTTATAAGCATGTTATGGGACTGAAATGGATTTCACATCATTGTATGAACTCAAAATATATTTTAAAGACTGATGATGATATAGTAGTGAATGCTGATGAGTTAAAGAGATTTTTAGTGAGAAGATTGTCTCCTTGGGGGGCTAAAGGTTTGATAATGTGTAAAGTTGCAAAACATGCCTTAGCACAGAGGAGACAGAGTTCAAAATGGATGGTTACACTTGAGGAATACCCTATGCCGTTTTATCCTGACTATTGTCCAGGCTGGGCAATTTTATACTCCAGAGATGTTGTACCTCGTTTACTGGAAGCAGCACAAAACACACCTTACTTCTGGATAGATGATGTGCATATAACTGGAATTTTAGCACAGAAGATTGGGGTACCGAGAACAAGTATAACTAATTATGTATTGACTGAAAAAAAAGCTGAATTGTTAGTGAAATCAGGAAAGCAGAATATAGGTCGATTCCTGTTTGGGCCACCCAATTTAAAAGTTCAAAGAATGTCTGCATTATGGCGCGCCATATCTCAATCTTGTCATATTTAG
Protein
MTESQRPKERIEMITLHKILFVFCFLSLSLNYYISTKPMLDIENPTFSLIQPNETITPKFSDNYDQLIDIKKFSFKINPKPCKRYPEGFLLMIIVSSNPLNYENRLVIRKTWGQTDEATNIVFLVGETDNVTVSQKIQEESVTYGDIVQGNFKDAYHNMTYKHVMGLKWISHHCMNSKYILKTDDDIVVNADELKRFLVRRLSPWGAKGLIMCKVAKHALAQRRQSSKWMVTLEEYPMPFYPDYCPGWAILYSRDVVPRLLEAAQNTPYFWIDDVHITGILAQKIGVPRTSITNYVLTEKKAELLVKSGKQNIGRFLFGPPNLKVQRMSALWRAISQSCHI
Summary
Similarity
Belongs to the glycosyltransferase 31 family.
Feature
chain Hexosyltransferase
Uniprot
G1UHW3
H9J529
A0A142I0M5
A0A2A4JLU4
A0A2H1X0S7
A0A0L7LSK8
+ More
E2B9U7 A0A0M8ZSG0 A0A2P8ZMZ6 A0A0C9R0Z2 A0A0L7R8K9 A0A026W539 A0A088AJ14 A0A2A3E6E1 A0A0J7L6P5 A0A154PNL0 A0A0T6BGP0 E2AN20 A0A195DI43 A0A195D3N8 A0A195EQW8 F4WKH2 A0A195BSL0 A0A158NQL7 A0A151XAW8 D7GXV7 A0A067QVM3 A0A2J7QPG1 A0A2P8XZD9 A0A0A9ZF59 V5GN24 A0A1B6JEJ3 A0A232F8C6 A0A139WK07 A0A1B6EMC1 A0A1W4WYY7 A0A2P6LIV0 A0A1B6EB26 A0A2J7PEK1 A0A084WEQ5 A0A1J1J7K3 A0A182GM22 A0A182PDR8 A0A023F1E7 A0A1Y1KYH1 A0A2K8JUZ5 A0A182VFV0 A0A1Q3FWG1 A0A182M3G7 A0A182THJ0 A0A182Y2R3 A0A182I1D5 A7UU23 A0A1Q3FW19 A0A182RQX3 A0A182SBG6 A0A182XP91 A0A1B0CLE9 A0A182JRX3 J9JSC1 A0A182PZW5 A0A087TH79 A0A2M4CV34 A0A1B0GQ78 A0A1E1X780 A0A2H8TR47 A0A182WJX0 A0A182NJU5 A0A2P8XT05 A0A1Z5L479 A0A2M4DIT7 A0A2M4DIT5 A0A2M4DIU9 A0A2M4DIT9 A0A293LFC1 A0A2M4AQP3 A0A182JFM5 A0A2M4A7X7 A0A2M4AAU6 A0A2M4AHS4 A0A2M4ACJ7 A0A1E1XJD6 T1K8T3 A0A1B0CKS7 A0A1L8E1L9 A0A1L8E1I1 A0A2M4BJL0 A0A2M4BJG9 A0A2S2NA04 A0A1L8E1I9 A0A1L8E1G8 A0A131XSL1 A0A2M4BIZ7 A0A2M4BIX9 A0A1L8E1H5
E2B9U7 A0A0M8ZSG0 A0A2P8ZMZ6 A0A0C9R0Z2 A0A0L7R8K9 A0A026W539 A0A088AJ14 A0A2A3E6E1 A0A0J7L6P5 A0A154PNL0 A0A0T6BGP0 E2AN20 A0A195DI43 A0A195D3N8 A0A195EQW8 F4WKH2 A0A195BSL0 A0A158NQL7 A0A151XAW8 D7GXV7 A0A067QVM3 A0A2J7QPG1 A0A2P8XZD9 A0A0A9ZF59 V5GN24 A0A1B6JEJ3 A0A232F8C6 A0A139WK07 A0A1B6EMC1 A0A1W4WYY7 A0A2P6LIV0 A0A1B6EB26 A0A2J7PEK1 A0A084WEQ5 A0A1J1J7K3 A0A182GM22 A0A182PDR8 A0A023F1E7 A0A1Y1KYH1 A0A2K8JUZ5 A0A182VFV0 A0A1Q3FWG1 A0A182M3G7 A0A182THJ0 A0A182Y2R3 A0A182I1D5 A7UU23 A0A1Q3FW19 A0A182RQX3 A0A182SBG6 A0A182XP91 A0A1B0CLE9 A0A182JRX3 J9JSC1 A0A182PZW5 A0A087TH79 A0A2M4CV34 A0A1B0GQ78 A0A1E1X780 A0A2H8TR47 A0A182WJX0 A0A182NJU5 A0A2P8XT05 A0A1Z5L479 A0A2M4DIT7 A0A2M4DIT5 A0A2M4DIU9 A0A2M4DIT9 A0A293LFC1 A0A2M4AQP3 A0A182JFM5 A0A2M4A7X7 A0A2M4AAU6 A0A2M4AHS4 A0A2M4ACJ7 A0A1E1XJD6 T1K8T3 A0A1B0CKS7 A0A1L8E1L9 A0A1L8E1I1 A0A2M4BJL0 A0A2M4BJG9 A0A2S2NA04 A0A1L8E1I9 A0A1L8E1G8 A0A131XSL1 A0A2M4BIZ7 A0A2M4BIX9 A0A1L8E1H5
EC Number
2.4.1.-
Pubmed
EMBL
AB620070
BAK82122.1
BABH01038850
KR362875
AMR34864.1
NWSH01001042
+ More
PCG72951.1 ODYU01012470 SOQ58796.1 JTDY01000230 KOB78166.1 GL446605 EFN87534.1 KQ435922 KOX68589.1 PYGN01000013 PSN57881.1 GBYB01000546 JAG70313.1 KQ414632 KOC67163.1 KK107405 QOIP01000014 EZA51195.1 RLU14921.1 KZ288372 PBC26729.1 LBMM01000511 KMQ98179.1 KQ434998 KZC13307.1 LJIG01000423 KRT86517.1 GL441034 EFN65178.1 KQ980824 KYN12507.1 KQ976885 KYN07525.1 KQ982021 KYN30571.1 GL888199 EGI65310.1 KQ976419 KYM89285.1 ADTU01023390 KQ982335 KYQ57532.1 GG694706 EFA13595.1 KK852891 KDR14239.1 NEVH01012091 PNF30470.1 PYGN01001127 PSN37368.1 GBHO01000510 GBHO01000504 GBRD01002426 GDHC01014766 JAG43094.1 JAG43100.1 JAG63395.1 JAQ03863.1 GALX01005509 JAB62957.1 GECU01031163 GECU01010056 GECU01008753 GECU01003507 JAS76543.1 JAS97650.1 JAS98953.1 JAT04200.1 NNAY01000777 OXU26557.1 KQ971334 KYB28262.1 GECZ01030692 JAS39077.1 MWRG01000024 PRD38483.1 GEDC01008248 GEDC01002191 JAS29050.1 JAS35107.1 NEVH01026094 PNF14756.1 ATLV01023244 KE525341 KFB48699.1 CVRI01000075 CRL08367.1 JXUM01073685 KQ562789 KXJ75126.1 GBBI01003365 JAC15347.1 GEZM01072342 JAV65551.1 KY031134 ATU82885.1 GFDL01003239 JAV31806.1 AXCM01005027 APCN01000053 AAAB01008960 EDO63801.1 GFDL01003241 JAV31804.1 AJWK01017181 AJWK01017182 AJWK01017183 ABLF02032829 AXCN02000524 KK115214 KFM64468.1 GGFL01005028 MBW69206.1 AJVK01035968 GFAC01004066 JAT95122.1 GFXV01004665 MBW16470.1 PYGN01001393 PSN35141.1 GFJQ02004763 JAW02207.1 GGFL01013292 MBW77470.1 GGFL01013293 MBW77471.1 GGFL01013295 MBW77473.1 GGFL01013294 MBW77472.1 GFWV01000665 MAA25395.1 GGFK01009794 MBW43115.1 AXCP01008135 GGFK01003576 MBW36897.1 GGFK01004590 MBW37911.1 GGFK01007024 MBW40345.1 GGFK01005168 MBW38489.1 GFAA01004195 JAT99239.1 CAEY01001881 AJWK01016600 GFDF01001505 JAV12579.1 GFDF01001506 JAV12578.1 GGFJ01004081 MBW53222.1 GGFJ01004078 MBW53219.1 GGMR01001432 MBY14051.1 GFDF01001503 JAV12581.1 GFDF01001501 JAV12583.1 GEFM01005503 JAP70293.1 GGFJ01003884 MBW53025.1 GGFJ01003885 MBW53026.1 GFDF01001504 JAV12580.1
PCG72951.1 ODYU01012470 SOQ58796.1 JTDY01000230 KOB78166.1 GL446605 EFN87534.1 KQ435922 KOX68589.1 PYGN01000013 PSN57881.1 GBYB01000546 JAG70313.1 KQ414632 KOC67163.1 KK107405 QOIP01000014 EZA51195.1 RLU14921.1 KZ288372 PBC26729.1 LBMM01000511 KMQ98179.1 KQ434998 KZC13307.1 LJIG01000423 KRT86517.1 GL441034 EFN65178.1 KQ980824 KYN12507.1 KQ976885 KYN07525.1 KQ982021 KYN30571.1 GL888199 EGI65310.1 KQ976419 KYM89285.1 ADTU01023390 KQ982335 KYQ57532.1 GG694706 EFA13595.1 KK852891 KDR14239.1 NEVH01012091 PNF30470.1 PYGN01001127 PSN37368.1 GBHO01000510 GBHO01000504 GBRD01002426 GDHC01014766 JAG43094.1 JAG43100.1 JAG63395.1 JAQ03863.1 GALX01005509 JAB62957.1 GECU01031163 GECU01010056 GECU01008753 GECU01003507 JAS76543.1 JAS97650.1 JAS98953.1 JAT04200.1 NNAY01000777 OXU26557.1 KQ971334 KYB28262.1 GECZ01030692 JAS39077.1 MWRG01000024 PRD38483.1 GEDC01008248 GEDC01002191 JAS29050.1 JAS35107.1 NEVH01026094 PNF14756.1 ATLV01023244 KE525341 KFB48699.1 CVRI01000075 CRL08367.1 JXUM01073685 KQ562789 KXJ75126.1 GBBI01003365 JAC15347.1 GEZM01072342 JAV65551.1 KY031134 ATU82885.1 GFDL01003239 JAV31806.1 AXCM01005027 APCN01000053 AAAB01008960 EDO63801.1 GFDL01003241 JAV31804.1 AJWK01017181 AJWK01017182 AJWK01017183 ABLF02032829 AXCN02000524 KK115214 KFM64468.1 GGFL01005028 MBW69206.1 AJVK01035968 GFAC01004066 JAT95122.1 GFXV01004665 MBW16470.1 PYGN01001393 PSN35141.1 GFJQ02004763 JAW02207.1 GGFL01013292 MBW77470.1 GGFL01013293 MBW77471.1 GGFL01013295 MBW77473.1 GGFL01013294 MBW77472.1 GFWV01000665 MAA25395.1 GGFK01009794 MBW43115.1 AXCP01008135 GGFK01003576 MBW36897.1 GGFK01004590 MBW37911.1 GGFK01007024 MBW40345.1 GGFK01005168 MBW38489.1 GFAA01004195 JAT99239.1 CAEY01001881 AJWK01016600 GFDF01001505 JAV12579.1 GFDF01001506 JAV12578.1 GGFJ01004081 MBW53222.1 GGFJ01004078 MBW53219.1 GGMR01001432 MBY14051.1 GFDF01001503 JAV12581.1 GFDF01001501 JAV12583.1 GEFM01005503 JAP70293.1 GGFJ01003884 MBW53025.1 GGFJ01003885 MBW53026.1 GFDF01001504 JAV12580.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000008237
UP000053105
UP000245037
+ More
UP000053825 UP000053097 UP000279307 UP000005203 UP000242457 UP000036403 UP000076502 UP000000311 UP000078492 UP000078542 UP000078541 UP000007755 UP000078540 UP000005205 UP000075809 UP000007266 UP000027135 UP000235965 UP000215335 UP000192223 UP000030765 UP000183832 UP000069940 UP000249989 UP000075885 UP000075903 UP000075883 UP000075902 UP000076408 UP000075840 UP000007062 UP000075900 UP000075901 UP000076407 UP000092461 UP000075881 UP000007819 UP000075886 UP000054359 UP000092462 UP000075920 UP000075884 UP000075880 UP000015104
UP000053825 UP000053097 UP000279307 UP000005203 UP000242457 UP000036403 UP000076502 UP000000311 UP000078492 UP000078542 UP000078541 UP000007755 UP000078540 UP000005205 UP000075809 UP000007266 UP000027135 UP000235965 UP000215335 UP000192223 UP000030765 UP000183832 UP000069940 UP000249989 UP000075885 UP000075903 UP000075883 UP000075902 UP000076408 UP000075840 UP000007062 UP000075900 UP000075901 UP000076407 UP000092461 UP000075881 UP000007819 UP000075886 UP000054359 UP000092462 UP000075920 UP000075884 UP000075880 UP000015104
Pfam
PF01762 Galactosyl_T
SUPFAM
SSF53448
SSF53448
ProteinModelPortal
G1UHW3
H9J529
A0A142I0M5
A0A2A4JLU4
A0A2H1X0S7
A0A0L7LSK8
+ More
E2B9U7 A0A0M8ZSG0 A0A2P8ZMZ6 A0A0C9R0Z2 A0A0L7R8K9 A0A026W539 A0A088AJ14 A0A2A3E6E1 A0A0J7L6P5 A0A154PNL0 A0A0T6BGP0 E2AN20 A0A195DI43 A0A195D3N8 A0A195EQW8 F4WKH2 A0A195BSL0 A0A158NQL7 A0A151XAW8 D7GXV7 A0A067QVM3 A0A2J7QPG1 A0A2P8XZD9 A0A0A9ZF59 V5GN24 A0A1B6JEJ3 A0A232F8C6 A0A139WK07 A0A1B6EMC1 A0A1W4WYY7 A0A2P6LIV0 A0A1B6EB26 A0A2J7PEK1 A0A084WEQ5 A0A1J1J7K3 A0A182GM22 A0A182PDR8 A0A023F1E7 A0A1Y1KYH1 A0A2K8JUZ5 A0A182VFV0 A0A1Q3FWG1 A0A182M3G7 A0A182THJ0 A0A182Y2R3 A0A182I1D5 A7UU23 A0A1Q3FW19 A0A182RQX3 A0A182SBG6 A0A182XP91 A0A1B0CLE9 A0A182JRX3 J9JSC1 A0A182PZW5 A0A087TH79 A0A2M4CV34 A0A1B0GQ78 A0A1E1X780 A0A2H8TR47 A0A182WJX0 A0A182NJU5 A0A2P8XT05 A0A1Z5L479 A0A2M4DIT7 A0A2M4DIT5 A0A2M4DIU9 A0A2M4DIT9 A0A293LFC1 A0A2M4AQP3 A0A182JFM5 A0A2M4A7X7 A0A2M4AAU6 A0A2M4AHS4 A0A2M4ACJ7 A0A1E1XJD6 T1K8T3 A0A1B0CKS7 A0A1L8E1L9 A0A1L8E1I1 A0A2M4BJL0 A0A2M4BJG9 A0A2S2NA04 A0A1L8E1I9 A0A1L8E1G8 A0A131XSL1 A0A2M4BIZ7 A0A2M4BIX9 A0A1L8E1H5
E2B9U7 A0A0M8ZSG0 A0A2P8ZMZ6 A0A0C9R0Z2 A0A0L7R8K9 A0A026W539 A0A088AJ14 A0A2A3E6E1 A0A0J7L6P5 A0A154PNL0 A0A0T6BGP0 E2AN20 A0A195DI43 A0A195D3N8 A0A195EQW8 F4WKH2 A0A195BSL0 A0A158NQL7 A0A151XAW8 D7GXV7 A0A067QVM3 A0A2J7QPG1 A0A2P8XZD9 A0A0A9ZF59 V5GN24 A0A1B6JEJ3 A0A232F8C6 A0A139WK07 A0A1B6EMC1 A0A1W4WYY7 A0A2P6LIV0 A0A1B6EB26 A0A2J7PEK1 A0A084WEQ5 A0A1J1J7K3 A0A182GM22 A0A182PDR8 A0A023F1E7 A0A1Y1KYH1 A0A2K8JUZ5 A0A182VFV0 A0A1Q3FWG1 A0A182M3G7 A0A182THJ0 A0A182Y2R3 A0A182I1D5 A7UU23 A0A1Q3FW19 A0A182RQX3 A0A182SBG6 A0A182XP91 A0A1B0CLE9 A0A182JRX3 J9JSC1 A0A182PZW5 A0A087TH79 A0A2M4CV34 A0A1B0GQ78 A0A1E1X780 A0A2H8TR47 A0A182WJX0 A0A182NJU5 A0A2P8XT05 A0A1Z5L479 A0A2M4DIT7 A0A2M4DIT5 A0A2M4DIU9 A0A2M4DIT9 A0A293LFC1 A0A2M4AQP3 A0A182JFM5 A0A2M4A7X7 A0A2M4AAU6 A0A2M4AHS4 A0A2M4ACJ7 A0A1E1XJD6 T1K8T3 A0A1B0CKS7 A0A1L8E1L9 A0A1L8E1I1 A0A2M4BJL0 A0A2M4BJG9 A0A2S2NA04 A0A1L8E1I9 A0A1L8E1G8 A0A131XSL1 A0A2M4BIZ7 A0A2M4BIX9 A0A1L8E1H5
Ontologies
GO
PANTHER
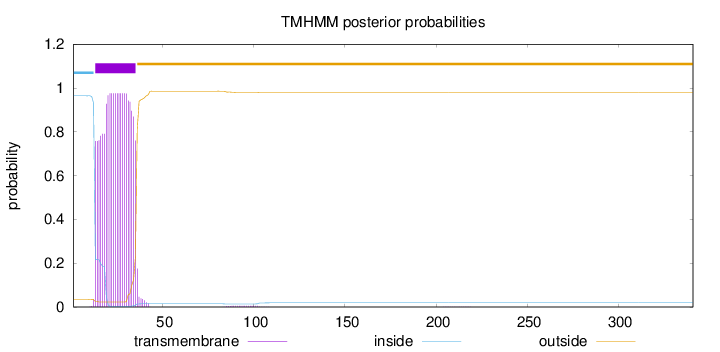
Topology
Subcellular location
Golgi apparatus membrane
Length:
341
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.2071099999999
Exp number, first 60 AAs:
21.10188
Total prob of N-in:
0.96451
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 341
Population Genetic Test Statistics
Pi
258.438571
Theta
213.581242
Tajima's D
0.641199
CLR
0
CSRT
0.56097195140243
Interpretation
Uncertain
