Gene
KWMTBOMO16100
Pre Gene Modal
BGIBMGA004624
Annotation
PREDICTED:_glycine_receptor_subunit_alpha-2-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.668
Sequence
CDS
ATGGCGGCTGGAAAGCTGCTGTGGCTTACGACTGCTTTGCTACGACTTTGGCTGGGAGCTACCGACTTGTGTTACGCGCAGTCAACACTCCTGACCTTCCAAGACCTCCTTCCTGATGACCCCCGACTGTACGACAAGATGCGCCCTCCTAAGATCAACGACCAGCCCACCACCGTGAACTTCCACGTCACAGTAATGGGTTTAGACTCCATAGACGAGAACTCAATGACCTACGCTGCGGATATATTCTTCGCACAGACGTGGAAAGATTTTCGTCTACGCTTGCCAGAGAACATGACTTCTGAATATCGGCTGTTAGAAGTGGACTGGCTGAAACACATGTGGCGCCCAGACTCGTTCTTCAAGAACGCCAAGTCGGTGACGTTTCAGACGATGACCATACCGAACCACTACGTGTGGCTGTACAAGGACAAGACCATACTGTACATGGTGAAGCTGACGTTGAGGCTGTCTTGCGCTATGAACTTCCTCATTTACCCTCACGATACGCAGGAGTGCAAACTGCAGATGGAGAGTCAACGTCTCATTCCTGTCGTTTAA
Protein
MAAGKLLWLTTALLRLWLGATDLCYAQSTLLTFQDLLPDDPRLYDKMRPPKINDQPTTVNFHVTVMGLDSIDENSMTYAADIFFAQTWKDFRLRLPENMTSEYRLLEVDWLKHMWRPDSFFKNAKSVTFQTMTIPNHYVWLYKDKTILYMVKLTLRLSCAMNFLIYPHDTQECKLQMESQRLIPVV
Summary
Similarity
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Belongs to the small heat shock protein (HSP20) family.
Belongs to the small heat shock protein (HSP20) family.
Uniprot
H9J534
A0A0N1PIU4
A0A212FPH0
A0A2H1W8M8
A0A194QJK2
A0A2W1BQQ4
+ More
A0A2A4JHW4 A0A182QRK7 A0A182N5Z1 A0A182SUI5 A0A182JAP1 A0A084VRZ0 A0A182YJS5 A0A182R1K3 A0A1B6C9S2 A0A182FQJ1 A0A182K0E9 A0A182V8X8 B0X4C3 A0A182KM32 A0A182XBK0 Q7PYF3 A0A182I0D9 A0A182TY19 A0A182GB84 A0A182LT77 A0A1S4FVK1 A0A0J7KED0 A0A182VQZ5 Q16MN1 W5J3F3 A0A2J7QMJ8 A0A182PC39 F4X3L1 A0A158NZZ9 A0A1B0EU65 Q0GQR2 A0A139WID5 A0A1B1QGE4 A8DMU7 A0A310SGT2 E2BB08 A0A139WIM3 D3UAF0 A0A0C9R0N0 A0A195CQM4 A0A026VTT1 A0A336L4G9 K7IXV1 E2ACX9 A0A195BN15 A0A336L8K3 A0A2R7WEV2 A0A3L8DW80 E0VZ48 A0A0R1EFH3 A0A2S2NHC1 A0A2S2Q405 W8BRS1 A0A0K8UTP5 A0A034WD30 A0A1J1IF71 B4JT27 Q9VDU9 Q6TF31 Q6TF30 B3P093 B4QSV3 B4IHT1 B4PNJ0 A0A1W4W826 A0A0M5J2B8 B4M4Q2 Q293L8 B3LXQ8 A0A3B0JRW1 B4K4T7 B4NF02 A0A2A3ESK0 B4GLY0 A0A1B0FHR8 A1KYB5 A1KYB3 A0A1A9WRN1 A0A0L0CP68 A0A1I8MBK0 A0A1A9VMB7 A0A1A9ZPQ1 A1KYB4 A0A1I8NXM5 A0A0M9A1N8 A0A1A9YF81 A0A154NWV9 A0A1D2MVS3 N6TWP0 A0A151X4E1 A0A1B3P9B7 A0A0L7RHJ3 A0A3R7MHZ6 A0A195E076
A0A2A4JHW4 A0A182QRK7 A0A182N5Z1 A0A182SUI5 A0A182JAP1 A0A084VRZ0 A0A182YJS5 A0A182R1K3 A0A1B6C9S2 A0A182FQJ1 A0A182K0E9 A0A182V8X8 B0X4C3 A0A182KM32 A0A182XBK0 Q7PYF3 A0A182I0D9 A0A182TY19 A0A182GB84 A0A182LT77 A0A1S4FVK1 A0A0J7KED0 A0A182VQZ5 Q16MN1 W5J3F3 A0A2J7QMJ8 A0A182PC39 F4X3L1 A0A158NZZ9 A0A1B0EU65 Q0GQR2 A0A139WID5 A0A1B1QGE4 A8DMU7 A0A310SGT2 E2BB08 A0A139WIM3 D3UAF0 A0A0C9R0N0 A0A195CQM4 A0A026VTT1 A0A336L4G9 K7IXV1 E2ACX9 A0A195BN15 A0A336L8K3 A0A2R7WEV2 A0A3L8DW80 E0VZ48 A0A0R1EFH3 A0A2S2NHC1 A0A2S2Q405 W8BRS1 A0A0K8UTP5 A0A034WD30 A0A1J1IF71 B4JT27 Q9VDU9 Q6TF31 Q6TF30 B3P093 B4QSV3 B4IHT1 B4PNJ0 A0A1W4W826 A0A0M5J2B8 B4M4Q2 Q293L8 B3LXQ8 A0A3B0JRW1 B4K4T7 B4NF02 A0A2A3ESK0 B4GLY0 A0A1B0FHR8 A1KYB5 A1KYB3 A0A1A9WRN1 A0A0L0CP68 A0A1I8MBK0 A0A1A9VMB7 A0A1A9ZPQ1 A1KYB4 A0A1I8NXM5 A0A0M9A1N8 A0A1A9YF81 A0A154NWV9 A0A1D2MVS3 N6TWP0 A0A151X4E1 A0A1B3P9B7 A0A0L7RHJ3 A0A3R7MHZ6 A0A195E076
Pubmed
19121390
26354079
22118469
28756777
24438588
25244985
+ More
20966253 12364791 14747013 17210077 26483478 17510324 20920257 23761445 21719571 21347285 16902773 18362917 19820115 17880682 20798317 20087392 24508170 20075255 30249741 20566863 17994087 17550304 24495485 25348373 10731132 12537568 12537572 12537573 12537574 11714703 11753412 16110336 17569856 17569867 26109357 26109356 16360945 15632085 23185243 18057021 26108605 25315136 27289101 23537049
20966253 12364791 14747013 17210077 26483478 17510324 20920257 23761445 21719571 21347285 16902773 18362917 19820115 17880682 20798317 20087392 24508170 20075255 30249741 20566863 17994087 17550304 24495485 25348373 10731132 12537568 12537572 12537573 12537574 11714703 11753412 16110336 17569856 17569867 26109357 26109356 16360945 15632085 23185243 18057021 26108605 25315136 27289101 23537049
EMBL
BABH01038851
BABH01038852
KQ460077
KPJ17892.1
AGBW02002687
OWR55627.1
+ More
ODYU01006917 SOQ49192.1 LC373508 KQ458671 BBD14178.1 KPJ05564.1 KZ150011 PZC75070.1 NWSH01001510 PCG71020.1 AXCN02000191 ATLV01015787 KE525036 KFB40734.1 GEDC01027061 JAS10237.1 DS232337 EDS40276.1 EDS40288.1 AAAB01008987 EAA01021.5 APCN01002072 JXUM01054215 JXUM01054216 KQ561810 KXJ77459.1 AXCM01007470 LBMM01008736 KMQ88662.1 CH477856 EAT35586.1 ADMH02002134 ETN58371.1 NEVH01013208 PNF29773.1 GL888624 EGI58808.1 ADTU01005179 ADTU01005180 AJWK01014063 AJWK01014064 AJWK01014065 AJWK01014066 DQ667187 ABG75739.1 KQ971338 KYB27770.1 KU532293 ANT80568.1 EF545124 ABU63602.1 KQ763877 OAD54660.1 GL446901 EFN87099.1 KYB27769.1 FJ851089 ACZ51421.1 GBYB01001545 JAG71312.1 KQ977394 KYN03006.1 KK107929 EZA47107.1 UFQS01001971 UFQT01001971 SSX12806.1 SSX32248.1 GL438602 EFN68709.1 KQ976440 KYM86574.1 SSX12805.1 SSX32247.1 KK854669 PTY17761.1 QOIP01000003 RLU24616.1 DS235849 EEB18654.1 CH892399 KRK05495.1 GGMR01003982 MBY16601.1 GGMS01003282 MBY72485.1 GAMC01006972 JAB99583.1 GDHF01022267 JAI30047.1 GAKP01006363 JAC52589.1 CVRI01000048 CRK98864.1 CH916373 EDV94917.1 AE014297 AF411340 AY049774 AF382403 AF435469 BT029033 AAF55691.1 AAL05873.1 AAL12210.1 AAL66188.1 AAL74413.1 ABJ16966.1 AY422812 AAR33080.1 AY422813 AAR33081.2 CH954181 EDV48467.1 CM000364 EDX12313.1 CH480840 EDW49448.1 CM000160 EDW96053.1 CP012526 ALC46948.1 CH940652 EDW59613.1 CM000070 EAL29196.1 KRT01375.1 CH902617 EDV41715.1 KPU79308.1 OUUW01000008 SPP83733.1 CH933806 EDW16090.1 KRG01921.1 CH964251 EDW83377.1 KZ288189 PBC34677.1 CH479185 EDW38554.1 CCAG010015445 AY672641 AAV91002.1 AY672639 AAV91000.1 JRES01000097 KNC34130.1 AY672640 AAV91001.1 KQ435794 KOX73989.1 KQ434775 KZC04083.1 LJIJ01000465 ODM97183.1 APGK01049251 KB741146 KB631697 ENN73700.1 ERL85444.1 KQ982557 KYQ55088.1 KU716103 AOG14387.1 KQ414591 KOC70268.1 QCYY01003768 ROT62203.1 KQ979955 KYN18545.1
ODYU01006917 SOQ49192.1 LC373508 KQ458671 BBD14178.1 KPJ05564.1 KZ150011 PZC75070.1 NWSH01001510 PCG71020.1 AXCN02000191 ATLV01015787 KE525036 KFB40734.1 GEDC01027061 JAS10237.1 DS232337 EDS40276.1 EDS40288.1 AAAB01008987 EAA01021.5 APCN01002072 JXUM01054215 JXUM01054216 KQ561810 KXJ77459.1 AXCM01007470 LBMM01008736 KMQ88662.1 CH477856 EAT35586.1 ADMH02002134 ETN58371.1 NEVH01013208 PNF29773.1 GL888624 EGI58808.1 ADTU01005179 ADTU01005180 AJWK01014063 AJWK01014064 AJWK01014065 AJWK01014066 DQ667187 ABG75739.1 KQ971338 KYB27770.1 KU532293 ANT80568.1 EF545124 ABU63602.1 KQ763877 OAD54660.1 GL446901 EFN87099.1 KYB27769.1 FJ851089 ACZ51421.1 GBYB01001545 JAG71312.1 KQ977394 KYN03006.1 KK107929 EZA47107.1 UFQS01001971 UFQT01001971 SSX12806.1 SSX32248.1 GL438602 EFN68709.1 KQ976440 KYM86574.1 SSX12805.1 SSX32247.1 KK854669 PTY17761.1 QOIP01000003 RLU24616.1 DS235849 EEB18654.1 CH892399 KRK05495.1 GGMR01003982 MBY16601.1 GGMS01003282 MBY72485.1 GAMC01006972 JAB99583.1 GDHF01022267 JAI30047.1 GAKP01006363 JAC52589.1 CVRI01000048 CRK98864.1 CH916373 EDV94917.1 AE014297 AF411340 AY049774 AF382403 AF435469 BT029033 AAF55691.1 AAL05873.1 AAL12210.1 AAL66188.1 AAL74413.1 ABJ16966.1 AY422812 AAR33080.1 AY422813 AAR33081.2 CH954181 EDV48467.1 CM000364 EDX12313.1 CH480840 EDW49448.1 CM000160 EDW96053.1 CP012526 ALC46948.1 CH940652 EDW59613.1 CM000070 EAL29196.1 KRT01375.1 CH902617 EDV41715.1 KPU79308.1 OUUW01000008 SPP83733.1 CH933806 EDW16090.1 KRG01921.1 CH964251 EDW83377.1 KZ288189 PBC34677.1 CH479185 EDW38554.1 CCAG010015445 AY672641 AAV91002.1 AY672639 AAV91000.1 JRES01000097 KNC34130.1 AY672640 AAV91001.1 KQ435794 KOX73989.1 KQ434775 KZC04083.1 LJIJ01000465 ODM97183.1 APGK01049251 KB741146 KB631697 ENN73700.1 ERL85444.1 KQ982557 KYQ55088.1 KU716103 AOG14387.1 KQ414591 KOC70268.1 QCYY01003768 ROT62203.1 KQ979955 KYN18545.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000053268
UP000218220
UP000075886
+ More
UP000075884 UP000075901 UP000075880 UP000030765 UP000076408 UP000075900 UP000069272 UP000075881 UP000075903 UP000002320 UP000075882 UP000076407 UP000007062 UP000075840 UP000075902 UP000069940 UP000249989 UP000075883 UP000036403 UP000075920 UP000008820 UP000000673 UP000235965 UP000075885 UP000007755 UP000005205 UP000092461 UP000005203 UP000007266 UP000008237 UP000078542 UP000053097 UP000002358 UP000000311 UP000078540 UP000279307 UP000009046 UP000002282 UP000183832 UP000001070 UP000000803 UP000008711 UP000000304 UP000001292 UP000192221 UP000092553 UP000008792 UP000001819 UP000007801 UP000268350 UP000009192 UP000007798 UP000242457 UP000008744 UP000092444 UP000091820 UP000037069 UP000095301 UP000078200 UP000092445 UP000095300 UP000053105 UP000092443 UP000076502 UP000094527 UP000019118 UP000030742 UP000075809 UP000053825 UP000283509 UP000078492
UP000075884 UP000075901 UP000075880 UP000030765 UP000076408 UP000075900 UP000069272 UP000075881 UP000075903 UP000002320 UP000075882 UP000076407 UP000007062 UP000075840 UP000075902 UP000069940 UP000249989 UP000075883 UP000036403 UP000075920 UP000008820 UP000000673 UP000235965 UP000075885 UP000007755 UP000005205 UP000092461 UP000005203 UP000007266 UP000008237 UP000078542 UP000053097 UP000002358 UP000000311 UP000078540 UP000279307 UP000009046 UP000002282 UP000183832 UP000001070 UP000000803 UP000008711 UP000000304 UP000001292 UP000192221 UP000092553 UP000008792 UP000001819 UP000007801 UP000268350 UP000009192 UP000007798 UP000242457 UP000008744 UP000092444 UP000091820 UP000037069 UP000095301 UP000078200 UP000092445 UP000095300 UP000053105 UP000092443 UP000076502 UP000094527 UP000019118 UP000030742 UP000075809 UP000053825 UP000283509 UP000078492
Pfam
Interpro
IPR006201
Neur_channel
+ More
IPR018000 Neurotransmitter_ion_chnl_CS
IPR036719 Neuro-gated_channel_TM_sf
IPR036734 Neur_chan_lig-bd_sf
IPR006029 Neurotrans-gated_channel_TM
IPR006202 Neur_chan_lig-bd
IPR006028 GABAA/Glycine_rcpt
IPR013015 Laminin_IV_B
IPR008211 Laminin_N
IPR000742 EGF-like_dom
IPR002049 Laminin_EGF
IPR038684 Laminin_N_sf
IPR002068 A-crystallin/Hsp20_dom
IPR008978 HSP20-like_chaperone
IPR034907 NDK-like_dom
IPR036390 WH_DNA-bd_sf
IPR032054 Cdt1_C
IPR014939 CDT1_Gemini-bd-like
IPR006602 Uncharacterised_DM10
IPR036850 NDK-like_dom_sf
IPR037993 NDPk7B
IPR014722 Rib_L2_dom2
IPR041997 KOW_RPL6
IPR008991 Translation_prot_SH3-like_sf
IPR000915 60S_ribosomal_L6E
IPR018000 Neurotransmitter_ion_chnl_CS
IPR036719 Neuro-gated_channel_TM_sf
IPR036734 Neur_chan_lig-bd_sf
IPR006029 Neurotrans-gated_channel_TM
IPR006202 Neur_chan_lig-bd
IPR006028 GABAA/Glycine_rcpt
IPR013015 Laminin_IV_B
IPR008211 Laminin_N
IPR000742 EGF-like_dom
IPR002049 Laminin_EGF
IPR038684 Laminin_N_sf
IPR002068 A-crystallin/Hsp20_dom
IPR008978 HSP20-like_chaperone
IPR034907 NDK-like_dom
IPR036390 WH_DNA-bd_sf
IPR032054 Cdt1_C
IPR014939 CDT1_Gemini-bd-like
IPR006602 Uncharacterised_DM10
IPR036850 NDK-like_dom_sf
IPR037993 NDPk7B
IPR014722 Rib_L2_dom2
IPR041997 KOW_RPL6
IPR008991 Translation_prot_SH3-like_sf
IPR000915 60S_ribosomal_L6E
SUPFAM
ProteinModelPortal
H9J534
A0A0N1PIU4
A0A212FPH0
A0A2H1W8M8
A0A194QJK2
A0A2W1BQQ4
+ More
A0A2A4JHW4 A0A182QRK7 A0A182N5Z1 A0A182SUI5 A0A182JAP1 A0A084VRZ0 A0A182YJS5 A0A182R1K3 A0A1B6C9S2 A0A182FQJ1 A0A182K0E9 A0A182V8X8 B0X4C3 A0A182KM32 A0A182XBK0 Q7PYF3 A0A182I0D9 A0A182TY19 A0A182GB84 A0A182LT77 A0A1S4FVK1 A0A0J7KED0 A0A182VQZ5 Q16MN1 W5J3F3 A0A2J7QMJ8 A0A182PC39 F4X3L1 A0A158NZZ9 A0A1B0EU65 Q0GQR2 A0A139WID5 A0A1B1QGE4 A8DMU7 A0A310SGT2 E2BB08 A0A139WIM3 D3UAF0 A0A0C9R0N0 A0A195CQM4 A0A026VTT1 A0A336L4G9 K7IXV1 E2ACX9 A0A195BN15 A0A336L8K3 A0A2R7WEV2 A0A3L8DW80 E0VZ48 A0A0R1EFH3 A0A2S2NHC1 A0A2S2Q405 W8BRS1 A0A0K8UTP5 A0A034WD30 A0A1J1IF71 B4JT27 Q9VDU9 Q6TF31 Q6TF30 B3P093 B4QSV3 B4IHT1 B4PNJ0 A0A1W4W826 A0A0M5J2B8 B4M4Q2 Q293L8 B3LXQ8 A0A3B0JRW1 B4K4T7 B4NF02 A0A2A3ESK0 B4GLY0 A0A1B0FHR8 A1KYB5 A1KYB3 A0A1A9WRN1 A0A0L0CP68 A0A1I8MBK0 A0A1A9VMB7 A0A1A9ZPQ1 A1KYB4 A0A1I8NXM5 A0A0M9A1N8 A0A1A9YF81 A0A154NWV9 A0A1D2MVS3 N6TWP0 A0A151X4E1 A0A1B3P9B7 A0A0L7RHJ3 A0A3R7MHZ6 A0A195E076
A0A2A4JHW4 A0A182QRK7 A0A182N5Z1 A0A182SUI5 A0A182JAP1 A0A084VRZ0 A0A182YJS5 A0A182R1K3 A0A1B6C9S2 A0A182FQJ1 A0A182K0E9 A0A182V8X8 B0X4C3 A0A182KM32 A0A182XBK0 Q7PYF3 A0A182I0D9 A0A182TY19 A0A182GB84 A0A182LT77 A0A1S4FVK1 A0A0J7KED0 A0A182VQZ5 Q16MN1 W5J3F3 A0A2J7QMJ8 A0A182PC39 F4X3L1 A0A158NZZ9 A0A1B0EU65 Q0GQR2 A0A139WID5 A0A1B1QGE4 A8DMU7 A0A310SGT2 E2BB08 A0A139WIM3 D3UAF0 A0A0C9R0N0 A0A195CQM4 A0A026VTT1 A0A336L4G9 K7IXV1 E2ACX9 A0A195BN15 A0A336L8K3 A0A2R7WEV2 A0A3L8DW80 E0VZ48 A0A0R1EFH3 A0A2S2NHC1 A0A2S2Q405 W8BRS1 A0A0K8UTP5 A0A034WD30 A0A1J1IF71 B4JT27 Q9VDU9 Q6TF31 Q6TF30 B3P093 B4QSV3 B4IHT1 B4PNJ0 A0A1W4W826 A0A0M5J2B8 B4M4Q2 Q293L8 B3LXQ8 A0A3B0JRW1 B4K4T7 B4NF02 A0A2A3ESK0 B4GLY0 A0A1B0FHR8 A1KYB5 A1KYB3 A0A1A9WRN1 A0A0L0CP68 A0A1I8MBK0 A0A1A9VMB7 A0A1A9ZPQ1 A1KYB4 A0A1I8NXM5 A0A0M9A1N8 A0A1A9YF81 A0A154NWV9 A0A1D2MVS3 N6TWP0 A0A151X4E1 A0A1B3P9B7 A0A0L7RHJ3 A0A3R7MHZ6 A0A195E076
PDB
5VDH
E-value=9.26187e-18,
Score=217
Ontologies
KEGG
GO
GO:0004888
GO:0016021
GO:0005230
GO:0005886
GO:1902476
GO:0034220
GO:0007268
GO:0050877
GO:0005254
GO:0007165
GO:0042391
GO:0043005
GO:0005887
GO:0045202
GO:0006811
GO:0019182
GO:0019183
GO:0030534
GO:0043052
GO:0045472
GO:0009636
GO:0006821
GO:0005840
GO:0006412
GO:0003735
GO:0006810
GO:0016020
GO:0001678
GO:0005536
GO:0046835
GO:0006281
GO:0006259
GO:0005216
PANTHER
Topology
SignalP
Position: 1 - 26,
Likelihood: 0.934315
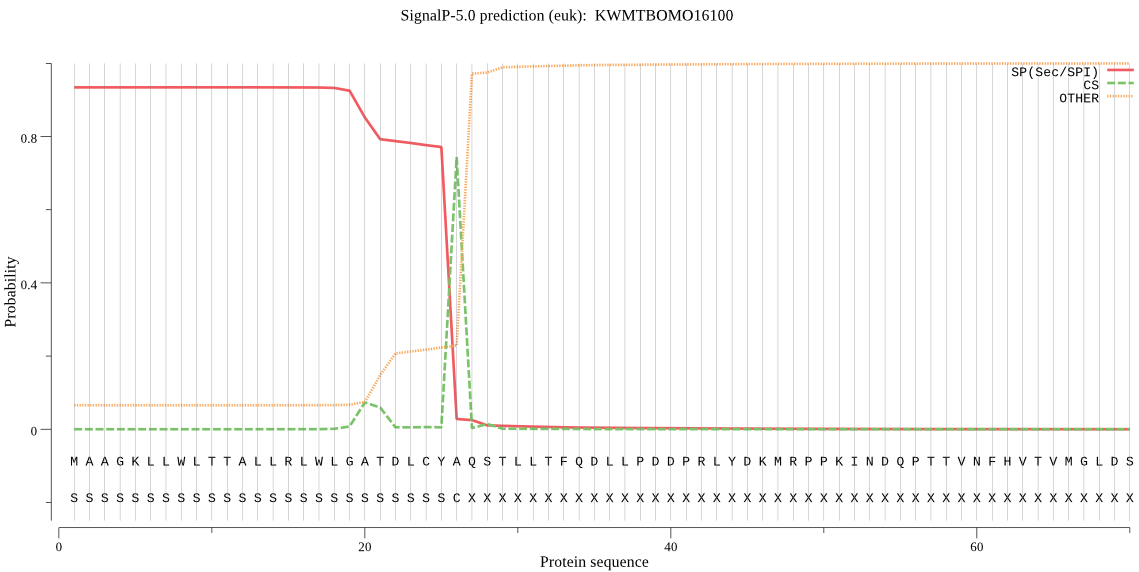
Length:
186
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.82962
Exp number, first 60 AAs:
0.54589
Total prob of N-in:
0.06521
outside
1 - 186

Population Genetic Test Statistics
Pi
218.1235
Theta
162.907612
Tajima's D
0.731503
CLR
1.449299
CSRT
0.579021048947553
Interpretation
Uncertain
