Gene
KWMTBOMO16099
Pre Gene Modal
BGIBMGA004625
Annotation
PREDICTED:_chloride_channel_protein_2_[Bombyx_mori]
Full name
Chloride channel protein
+ More
Chloride channel protein 2
Chloride channel protein 2
Alternative Name
Chloride channel-a
Location in the cell
PlasmaMembrane Reliability : 4.809
Sequence
CDS
ATGTATGGTCGCTATCAACGCGACCTCAGCGAAGCGGCAAGAGAAGAAGCAAGGAGACTCCGTAGACTGCGCAAGAAGAGACGCAGAGACGACAAACTCCGACAAAAGGAGCTGGCGGCTACCGGGAAACCGCGACCCAGGGGGAAGTTCTTTAAAGTTTTAGGCTACATATGGAGACACACGTTCGCTCGTTTAGGCGAAGACTGGATCTTTCTCGCCGTTCTCGGTATAATCACGGCTATACTTAATTTCGCTATGGACAGAGGGATCGCTATCTGCAATAATGCTCGCATGTGGATGTACAATGACCTGGCAACGTCCACGTTCAGTCAGTATGTGGCTTGGGTGTCTCTGCCCGTATGCCTGATACTATTCGCTGCTGGCTTCGTGCATATTGTTGCTGCACAGAGCATAGGTTCAGGCATACCGGAAATGAAGACGATTCTCCGAGGTGTCCACCTAAAAGAATATTTGACCTTCAGAGCACTCATATCCAAGCTCATCGGTCTCACTGCCACATTAGGATCTGGTCTTCCACTAGGGAAAGAAGGACCGTCAGTTCACATAGCCTCAATGGTGGCCACACTGCTCTCCAAGCTGGTGACCACCTTCCAAGGGATCTACAGCAACGAGTCTCGGAACAGCGAGATGTTGGCCGCAGCTTGTGCTGTCGGCGTGGCCTCGTGCTTTGCCGCACCTGTTGGCGGCGTGCTCTTCTCCATAGAAGTCACGACGACGTACTTCGCTGTGAGGAACTATTGGAGGGGATTCTTTGCGGCTTGCTGCGGGGCTATCATGTTCAGGCTTCTGTCGGTTTGGTCTGGCGCTATGCCGACTATCAAACCTCTGTTCCCAACGAACCTACCGCAGGACTTTCCTTTCGACCCCCAGGAGTTTGCCGTCTTCGTTCTTCTTGGTGTGGTGTGCGGTCTCTTAGCCTCGTTATGGATCTGGTTGCATCGGCAGTACGTTTTGTTTATGCGGAACACAAAAACCTTAAGCAACTTCTTGCAGAAGAACCGTTTCATATATCCCGGCTTCATAACTCTGGCCGTGATGTCGATACTGTTCCCGCCAGGCATCGGGAAGTACATGGCAGCAGTTCTAGGGAATCAGGAACAGGTGCTGTCCCTTTTCGCGAACTTCACTTGGTCCGATGAGTTGACCGCGGAACAAGCCTCAATAGTGTCTCATTGGAGGACACCCGAAGTTGAATATTGGGGGTGCCTCATCATCTTCTTCTTTTCTATTTTCTTCCTGAGTATGGTCTCTTGCACTCTGCCAGTTCCGGCGGGCATTTTCGTGCCGGCTTTCAAAATGGGAGCTTCGTTGGGACGGTTCACGGGCGAAGTGATGCACTACTTCTGTCCGCTCGGCGTGGCTTATGGAGGACACATCCAGAAGATTCTACCAGGAGGGTACGCGGTCGTCGGAGCCGCGGCCTTCACCGGCGCCGTCACTCACACTGTCTCCACCATTGTCATAGTCAGCGAAATGACCGGGCAGGTAACTCATCTCCTTGGCATAATGGCGGCGGTTTTGGCGGCAAATGCAGTAGCGGCTCTGCTTGGACCTTCCTGCTTCGACAGTATCATCCTCATCAAGAAGCTACCCTACCTGCCCGATCTGCTTTCCTCTGCTAGTCGTATGTATGACATTTGCGTGGAGGACTTCATGGTGCGTGAGGTCAAGTACATATGGAACAGAATGACCTTCCAACAACTGAAGGATCTCTTGAAAGAGAACAAGGCCATAAAGAGTTTCCCGCTGGTGGACAGTCCGGCGTCGATGGTGCTGCTGGGGTCCATCCACCGCTGGGAGCTGGTGCGCGTCATCGAGCGGCAGGTGGGGCGCTCGCGGCGGCTGCAGGTGGCCGCGCAGTGGCACCGCCGCGCGCTGCAGGACGAGGCCCGCGCCCGCACCAGGAGACCCTCCAGATTCGAGGTGACTCCTGCTCCGGACATGCTACACGCGCCGGGGACCGGCATGACGCGAGGGTCCTCGCTCACCACCAAGGACCAAGGAGGACTACTGCCATCTCCGGGTCAGCTGTTCCGGCCGAAGTCCATCCTGAAGAAGACGAACTCGTTCACGTTGACGCGCGGACTGGCCTCCCCGCCCCCGCAGGCGACCACCCCCTCCCCCGCGTCCTCCCCGGCGCAAGTGTACAGCACCGTCACCGGAGCCGAGACCAGGATCCGCGCGGCCTTCGAAGCCATCTTCAGGCGGTCGACGCTGCTGCAGGACGTGGAGGGAGGCCTCTCCGACCAGCTCACCCCCGGACTGCCGAGGAGTCCCTCCATCAACAAGAAAGTTCAACTGCCCCGCGAGCGAGTCTGCGACATGTCCCCCGAGGACCAGAAAGCCTGGGAGCAACTAGAGATGGCTAAAGAAATTGACTTCGATCGGATGCTTGGCATCGCCAGGAAACGAGATCTGCTGCCCGAGGACTCGGAGTACGAGGACGAGACGCTGTACATCTGCCACATCGACCCGGCGCCCTTCCAGCTGGTGGAGAGGACTTCTCTACTCAAGGTCCACTCACTGTTCTCCACGCTGGGGGTGAGCCGCGCTTACGTCACGGCCATCGGCAGACTCATCGGGGTCGTCGCGCTCAAAGAGCTCCGAAAAGCGATCGAAGACGTGAACAGCGGCGTGCTGACGGTCGCCACCCGGCCCTCGCCTCCCACCACCCCGCTGCCCACCATCATTAAGGTGCCCTCCACGCAGCCCGCACCGCCCTCCGACAGCGAGACCGCCAAACTAACGACGTCGGAGAAGACATAA
Protein
MYGRYQRDLSEAAREEARRLRRLRKKRRRDDKLRQKELAATGKPRPRGKFFKVLGYIWRHTFARLGEDWIFLAVLGIITAILNFAMDRGIAICNNARMWMYNDLATSTFSQYVAWVSLPVCLILFAAGFVHIVAAQSIGSGIPEMKTILRGVHLKEYLTFRALISKLIGLTATLGSGLPLGKEGPSVHIASMVATLLSKLVTTFQGIYSNESRNSEMLAAACAVGVASCFAAPVGGVLFSIEVTTTYFAVRNYWRGFFAACCGAIMFRLLSVWSGAMPTIKPLFPTNLPQDFPFDPQEFAVFVLLGVVCGLLASLWIWLHRQYVLFMRNTKTLSNFLQKNRFIYPGFITLAVMSILFPPGIGKYMAAVLGNQEQVLSLFANFTWSDELTAEQASIVSHWRTPEVEYWGCLIIFFFSIFFLSMVSCTLPVPAGIFVPAFKMGASLGRFTGEVMHYFCPLGVAYGGHIQKILPGGYAVVGAAAFTGAVTHTVSTIVIVSEMTGQVTHLLGIMAAVLAANAVAALLGPSCFDSIILIKKLPYLPDLLSSASRMYDICVEDFMVREVKYIWNRMTFQQLKDLLKENKAIKSFPLVDSPASMVLLGSIHRWELVRVIERQVGRSRRLQVAAQWHRRALQDEARARTRRPSRFEVTPAPDMLHAPGTGMTRGSSLTTKDQGGLLPSPGQLFRPKSILKKTNSFTLTRGLASPPPQATTPSPASSPAQVYSTVTGAETRIRAAFEAIFRRSTLLQDVEGGLSDQLTPGLPRSPSINKKVQLPRERVCDMSPEDQKAWEQLEMAKEIDFDRMLGIARKRDLLPEDSEYEDETLYICHIDPAPFQLVERTSLLKVHSLFSTLGVSRAYVTAIGRLIGVVALKELRKAIEDVNSGVLTVATRPSPPTTPLPTIIKVPSTQPAPPSDSETAKLTTSEKT
Summary
Description
Voltage-gated chloride channel. Chloride channels have several functions including the regulation of cell volume; membrane potential stabilization, signal transduction and transepithelial transport.
Miscellaneous
The CLC channel family contains both chloride channels and proton-coupled anion transporters that exchange chloride or another anion for protons. The absence of conserved gating glutamate residues is typical for family members that function as channels.
Similarity
Belongs to the chloride channel (TC 2.A.49) family.
Keywords
Alternative splicing
CBS domain
Chloride
Chloride channel
Complete proteome
Ion channel
Ion transport
Membrane
Reference proteome
Repeat
RNA editing
Transmembrane
Transmembrane helix
Transport
Voltage-gated channel
Feature
chain Chloride channel protein 2
splice variant In isoform G and isoform H.
splice variant In isoform G and isoform H.
Uniprot
A0A2W1BLB1
A0A194QKY5
A0A195BPF1
A0A158P2X7
A0A151WYL9
F4X527
+ More
E2AUK8 A0A0M9AA48 A0A154P3M6 A0A146M938 A0A026VZT3 E9ICR2 A0A0A9XWK7 A0A0C9RR94 A0A195DNB4 K7J6E3 A0A195EZZ8 A0A146LJA2 A0A0A9XR97 A0A1Q3G3D8 A0A1Q3G3D7 A0A1Q3G372 A0A1W4XVD9 A0A1W4XW78 A0A2H1W2K5 A0A0L7RD51 A0A023F429 A0A088AM85 A0A139WGD7 A0A310SMZ5 E0VHT5 A0A1I8NIP0 A0A1J1J6W0 A0A1I8NIP9 A0A0A1WTS8 T1I221 A0A0L0CK26 A0A034VCU2 W8B894 A0A034VA27 A0A182F979 A0A1I8P531 W8BZY0 A0A0K8TRF6 A0A2M4AGQ6 A0A2M4CG84 W5JW39 A0A1B6CT71 A0A1I8NIQ3 A0A0A1WL81 A0A0A1XPJ5 A0A0A1X0R7 A0A182RP26 W8BNB8 A0A034V7U2 A0A182P7R8 A0A034VCT9 A0A1I8P555 A0A182M8A1 A0A182JKU1 A0A182VYP3 Q9VGH7-4 A0A182XW91 Q9VGH7-6 A0A1W4W1I1 Q9VGH7-3 H1UUP0 A0A0R1E6S0 A0A1W4WDH5 A0A0R1E490 A0A1W4WC70 A0A182N708 A0A084VT41 A0A0R1E468 A0A182K0S4 A0A182HA86 A0A3B0KT60 A0A1B6FC04 I5APM4 I5APM2 A0A0Q9X818 A0A0Q9X638 B4N837 I5APM5 A0A1S4FC75 Q178C9 Q178D0 A0A0Q9X4L6 Q178C8 A0A0Q9W1C3 A0A0P9ARS4 A0A0P9A033 A0A182XHJ4 A0A0P8YFI5 F5HKH6 A0A1B6K060 A0A0Q9WDW6 A0A0Q9W1P6 F5HKH4
E2AUK8 A0A0M9AA48 A0A154P3M6 A0A146M938 A0A026VZT3 E9ICR2 A0A0A9XWK7 A0A0C9RR94 A0A195DNB4 K7J6E3 A0A195EZZ8 A0A146LJA2 A0A0A9XR97 A0A1Q3G3D8 A0A1Q3G3D7 A0A1Q3G372 A0A1W4XVD9 A0A1W4XW78 A0A2H1W2K5 A0A0L7RD51 A0A023F429 A0A088AM85 A0A139WGD7 A0A310SMZ5 E0VHT5 A0A1I8NIP0 A0A1J1J6W0 A0A1I8NIP9 A0A0A1WTS8 T1I221 A0A0L0CK26 A0A034VCU2 W8B894 A0A034VA27 A0A182F979 A0A1I8P531 W8BZY0 A0A0K8TRF6 A0A2M4AGQ6 A0A2M4CG84 W5JW39 A0A1B6CT71 A0A1I8NIQ3 A0A0A1WL81 A0A0A1XPJ5 A0A0A1X0R7 A0A182RP26 W8BNB8 A0A034V7U2 A0A182P7R8 A0A034VCT9 A0A1I8P555 A0A182M8A1 A0A182JKU1 A0A182VYP3 Q9VGH7-4 A0A182XW91 Q9VGH7-6 A0A1W4W1I1 Q9VGH7-3 H1UUP0 A0A0R1E6S0 A0A1W4WDH5 A0A0R1E490 A0A1W4WC70 A0A182N708 A0A084VT41 A0A0R1E468 A0A182K0S4 A0A182HA86 A0A3B0KT60 A0A1B6FC04 I5APM4 I5APM2 A0A0Q9X818 A0A0Q9X638 B4N837 I5APM5 A0A1S4FC75 Q178C9 Q178D0 A0A0Q9X4L6 Q178C8 A0A0Q9W1C3 A0A0P9ARS4 A0A0P9A033 A0A182XHJ4 A0A0P8YFI5 F5HKH6 A0A1B6K060 A0A0Q9WDW6 A0A0Q9W1P6 F5HKH4
Pubmed
28756777
26354079
21347285
21719571
20798317
26823975
+ More
24508170 30249741 21282665 25401762 20075255 25474469 18362917 19820115 20566863 25315136 25830018 26108605 25348373 24495485 26369729 20920257 23761445 10731132 12537572 12537569 15501232 17018572 17601953 25244985 17994087 17550304 24438588 26483478 15632085 17510324 12364791 14747013 17210077
24508170 30249741 21282665 25401762 20075255 25474469 18362917 19820115 20566863 25315136 25830018 26108605 25348373 24495485 26369729 20920257 23761445 10731132 12537572 12537569 15501232 17018572 17601953 25244985 17994087 17550304 24438588 26483478 15632085 17510324 12364791 14747013 17210077
EMBL
KZ150011
PZC75071.1
KQ458671
KPJ05565.1
KQ976432
KYM87521.1
+ More
ADTU01007642 ADTU01007643 KQ982649 KYQ52973.1 GL888680 EGI58484.1 GL442829 EFN62902.1 KQ435711 KOX79606.1 KQ434796 KZC05838.1 GDHC01003333 GDHC01002890 JAQ15296.1 JAQ15739.1 KK107538 QOIP01000005 EZA49145.1 RLU22975.1 GL762354 EFZ21619.1 GBHO01020341 JAG23263.1 GBYB01009821 JAG79588.1 KQ980713 KYN14395.1 KQ981905 KYN33449.1 GDHC01011244 JAQ07385.1 GBHO01020342 JAG23262.1 GFDL01000727 JAV34318.1 GFDL01000731 JAV34314.1 GFDL01000839 JAV34206.1 ODYU01005901 SOQ47267.1 KQ414615 KOC68686.1 GBBI01002501 JAC16211.1 KQ971345 KYB27012.1 KQ762882 OAD55366.1 DS235171 EEB12858.1 CVRI01000074 CRL08157.1 GBXI01011813 JAD02479.1 ACPB03006207 JRES01000301 KNC32600.1 GAKP01019584 JAC39368.1 GAMC01011693 JAB94862.1 GAKP01019588 JAC39364.1 GAMC01011691 JAB94864.1 GDAI01000882 JAI16721.1 GGFK01006652 MBW39973.1 GGFL01000067 MBW64245.1 ADMH02000272 ETN67164.1 GEDC01020875 JAS16423.1 GBXI01014670 JAC99621.1 GBXI01001799 JAD12493.1 GBXI01009957 JAD04335.1 GAMC01011694 JAB94861.1 GAKP01019586 JAC39366.1 GAKP01019589 JAC39363.1 AXCM01005921 AE014297 AY119539 BT009973 BT030454 AAF54702.3 AAM50193.1 AAQ22442.1 BT133125 AEY64293.1 CM000160 KRK03891.1 KRK03897.1 ATLV01016166 KE525060 KFB41135.1 KRK03893.1 JXUM01030864 KQ560899 KXJ80428.1 OUUW01000014 SPP88421.1 GECZ01022068 JAS47701.1 CM000070 EIM52909.1 EIM52907.1 CH933806 KRG00862.1 KRG00852.1 CH964232 EDW81288.2 EIM52910.2 CH477366 EAT42524.1 EAT42523.1 KRG00858.1 EAT42525.1 CH940652 KRF78876.1 CH902617 KPU80158.1 KPU80163.1 KPU80155.1 AAAB01008859 EGK96727.1 GECU01023788 GECU01002872 JAS83918.1 JAT04835.1 KRF78869.1 KRF78866.1 EGK96728.1
ADTU01007642 ADTU01007643 KQ982649 KYQ52973.1 GL888680 EGI58484.1 GL442829 EFN62902.1 KQ435711 KOX79606.1 KQ434796 KZC05838.1 GDHC01003333 GDHC01002890 JAQ15296.1 JAQ15739.1 KK107538 QOIP01000005 EZA49145.1 RLU22975.1 GL762354 EFZ21619.1 GBHO01020341 JAG23263.1 GBYB01009821 JAG79588.1 KQ980713 KYN14395.1 KQ981905 KYN33449.1 GDHC01011244 JAQ07385.1 GBHO01020342 JAG23262.1 GFDL01000727 JAV34318.1 GFDL01000731 JAV34314.1 GFDL01000839 JAV34206.1 ODYU01005901 SOQ47267.1 KQ414615 KOC68686.1 GBBI01002501 JAC16211.1 KQ971345 KYB27012.1 KQ762882 OAD55366.1 DS235171 EEB12858.1 CVRI01000074 CRL08157.1 GBXI01011813 JAD02479.1 ACPB03006207 JRES01000301 KNC32600.1 GAKP01019584 JAC39368.1 GAMC01011693 JAB94862.1 GAKP01019588 JAC39364.1 GAMC01011691 JAB94864.1 GDAI01000882 JAI16721.1 GGFK01006652 MBW39973.1 GGFL01000067 MBW64245.1 ADMH02000272 ETN67164.1 GEDC01020875 JAS16423.1 GBXI01014670 JAC99621.1 GBXI01001799 JAD12493.1 GBXI01009957 JAD04335.1 GAMC01011694 JAB94861.1 GAKP01019586 JAC39366.1 GAKP01019589 JAC39363.1 AXCM01005921 AE014297 AY119539 BT009973 BT030454 AAF54702.3 AAM50193.1 AAQ22442.1 BT133125 AEY64293.1 CM000160 KRK03891.1 KRK03897.1 ATLV01016166 KE525060 KFB41135.1 KRK03893.1 JXUM01030864 KQ560899 KXJ80428.1 OUUW01000014 SPP88421.1 GECZ01022068 JAS47701.1 CM000070 EIM52909.1 EIM52907.1 CH933806 KRG00862.1 KRG00852.1 CH964232 EDW81288.2 EIM52910.2 CH477366 EAT42524.1 EAT42523.1 KRG00858.1 EAT42525.1 CH940652 KRF78876.1 CH902617 KPU80158.1 KPU80163.1 KPU80155.1 AAAB01008859 EGK96727.1 GECU01023788 GECU01002872 JAS83918.1 JAT04835.1 KRF78869.1 KRF78866.1 EGK96728.1
Proteomes
UP000053268
UP000078540
UP000005205
UP000075809
UP000007755
UP000000311
+ More
UP000053105 UP000076502 UP000053097 UP000279307 UP000078492 UP000002358 UP000078541 UP000192223 UP000053825 UP000005203 UP000007266 UP000009046 UP000095301 UP000183832 UP000015103 UP000037069 UP000069272 UP000095300 UP000000673 UP000075900 UP000075885 UP000075883 UP000075880 UP000075920 UP000000803 UP000076408 UP000192221 UP000002282 UP000075884 UP000030765 UP000075881 UP000069940 UP000249989 UP000268350 UP000001819 UP000009192 UP000007798 UP000008820 UP000008792 UP000007801 UP000076407 UP000007062
UP000053105 UP000076502 UP000053097 UP000279307 UP000078492 UP000002358 UP000078541 UP000192223 UP000053825 UP000005203 UP000007266 UP000009046 UP000095301 UP000183832 UP000015103 UP000037069 UP000069272 UP000095300 UP000000673 UP000075900 UP000075885 UP000075883 UP000075880 UP000075920 UP000000803 UP000076408 UP000192221 UP000002282 UP000075884 UP000030765 UP000075881 UP000069940 UP000249989 UP000268350 UP000001819 UP000009192 UP000007798 UP000008820 UP000008792 UP000007801 UP000076407 UP000007062
SUPFAM
SSF81340
SSF81340
Gene 3D
ProteinModelPortal
A0A2W1BLB1
A0A194QKY5
A0A195BPF1
A0A158P2X7
A0A151WYL9
F4X527
+ More
E2AUK8 A0A0M9AA48 A0A154P3M6 A0A146M938 A0A026VZT3 E9ICR2 A0A0A9XWK7 A0A0C9RR94 A0A195DNB4 K7J6E3 A0A195EZZ8 A0A146LJA2 A0A0A9XR97 A0A1Q3G3D8 A0A1Q3G3D7 A0A1Q3G372 A0A1W4XVD9 A0A1W4XW78 A0A2H1W2K5 A0A0L7RD51 A0A023F429 A0A088AM85 A0A139WGD7 A0A310SMZ5 E0VHT5 A0A1I8NIP0 A0A1J1J6W0 A0A1I8NIP9 A0A0A1WTS8 T1I221 A0A0L0CK26 A0A034VCU2 W8B894 A0A034VA27 A0A182F979 A0A1I8P531 W8BZY0 A0A0K8TRF6 A0A2M4AGQ6 A0A2M4CG84 W5JW39 A0A1B6CT71 A0A1I8NIQ3 A0A0A1WL81 A0A0A1XPJ5 A0A0A1X0R7 A0A182RP26 W8BNB8 A0A034V7U2 A0A182P7R8 A0A034VCT9 A0A1I8P555 A0A182M8A1 A0A182JKU1 A0A182VYP3 Q9VGH7-4 A0A182XW91 Q9VGH7-6 A0A1W4W1I1 Q9VGH7-3 H1UUP0 A0A0R1E6S0 A0A1W4WDH5 A0A0R1E490 A0A1W4WC70 A0A182N708 A0A084VT41 A0A0R1E468 A0A182K0S4 A0A182HA86 A0A3B0KT60 A0A1B6FC04 I5APM4 I5APM2 A0A0Q9X818 A0A0Q9X638 B4N837 I5APM5 A0A1S4FC75 Q178C9 Q178D0 A0A0Q9X4L6 Q178C8 A0A0Q9W1C3 A0A0P9ARS4 A0A0P9A033 A0A182XHJ4 A0A0P8YFI5 F5HKH6 A0A1B6K060 A0A0Q9WDW6 A0A0Q9W1P6 F5HKH4
E2AUK8 A0A0M9AA48 A0A154P3M6 A0A146M938 A0A026VZT3 E9ICR2 A0A0A9XWK7 A0A0C9RR94 A0A195DNB4 K7J6E3 A0A195EZZ8 A0A146LJA2 A0A0A9XR97 A0A1Q3G3D8 A0A1Q3G3D7 A0A1Q3G372 A0A1W4XVD9 A0A1W4XW78 A0A2H1W2K5 A0A0L7RD51 A0A023F429 A0A088AM85 A0A139WGD7 A0A310SMZ5 E0VHT5 A0A1I8NIP0 A0A1J1J6W0 A0A1I8NIP9 A0A0A1WTS8 T1I221 A0A0L0CK26 A0A034VCU2 W8B894 A0A034VA27 A0A182F979 A0A1I8P531 W8BZY0 A0A0K8TRF6 A0A2M4AGQ6 A0A2M4CG84 W5JW39 A0A1B6CT71 A0A1I8NIQ3 A0A0A1WL81 A0A0A1XPJ5 A0A0A1X0R7 A0A182RP26 W8BNB8 A0A034V7U2 A0A182P7R8 A0A034VCT9 A0A1I8P555 A0A182M8A1 A0A182JKU1 A0A182VYP3 Q9VGH7-4 A0A182XW91 Q9VGH7-6 A0A1W4W1I1 Q9VGH7-3 H1UUP0 A0A0R1E6S0 A0A1W4WDH5 A0A0R1E490 A0A1W4WC70 A0A182N708 A0A084VT41 A0A0R1E468 A0A182K0S4 A0A182HA86 A0A3B0KT60 A0A1B6FC04 I5APM4 I5APM2 A0A0Q9X818 A0A0Q9X638 B4N837 I5APM5 A0A1S4FC75 Q178C9 Q178D0 A0A0Q9X4L6 Q178C8 A0A0Q9W1C3 A0A0P9ARS4 A0A0P9A033 A0A182XHJ4 A0A0P8YFI5 F5HKH6 A0A1B6K060 A0A0Q9WDW6 A0A0Q9W1P6 F5HKH4
PDB
6QVU
E-value=3.08658e-126,
Score=1160
Ontologies
GO
Topology
Subcellular location
Membrane
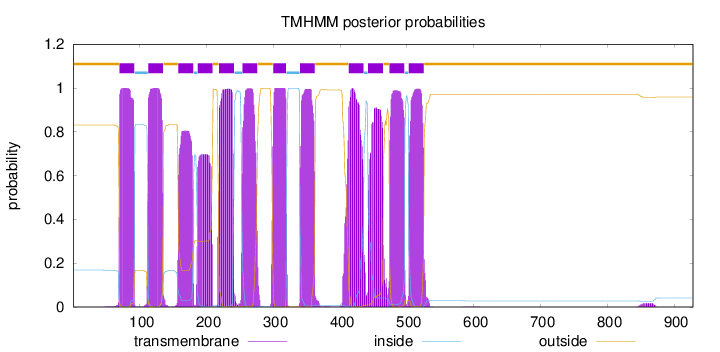
Length:
928
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
257.955299999999
Exp number, first 60 AAs:
0.01911
Total prob of N-in:
0.16854
outside
1 - 69
TMhelix
70 - 92
inside
93 - 112
TMhelix
113 - 135
outside
136 - 157
TMhelix
158 - 180
inside
181 - 186
TMhelix
187 - 209
outside
210 - 218
TMhelix
219 - 241
inside
242 - 253
TMhelix
254 - 276
outside
277 - 299
TMhelix
300 - 319
inside
320 - 339
TMhelix
340 - 362
outside
363 - 412
TMhelix
413 - 435
inside
436 - 441
TMhelix
442 - 464
outside
465 - 473
TMhelix
474 - 496
inside
497 - 502
TMhelix
503 - 525
outside
526 - 928
Population Genetic Test Statistics
Pi
280.72387
Theta
170.58651
Tajima's D
1.920429
CLR
0.21461
CSRT
0.873806309684516
Interpretation
Uncertain
