Gene
KWMTBOMO16098
Pre Gene Modal
BGIBMGA004618
Annotation
PREDICTED:_inhibin_beta_A_chain-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.35
Sequence
CDS
ATGATCGCGTGGAGCACAAGCGCGTTAGTGTTCATTCTAGTCGGTAATTTTTTAAGTTTGGATGGCGAGATGCTGGGCAGCTGCGCCAATTGTGCCAACGCGGACCTCAGTGTCACCCAGCTGACGGAGGCGCGGATCGAGTTCGTCAAGCAGCAGATATTGAAGAAGCTCCGCATGACCAAGAAGCCGGCTGTCGCGCTGCCCGTCAATAGCTTACCGATGCCGGTGGCGCAAGGACAGACCCTCAGTGAAGGCACTGATAAGCCCCCCGAGTTTGACGACTATTATGGCAGAACCGAACAAAAGATCATCTTCCCTGTCGAGGGAGAATGCTTAACGTCAGGCCGGTACCCATTCAAGTGTTTGCAGTTTGAGTTACCTCCGGACGTGGAACCTGAAGAAGTATCTGTGGTTGAACTTTGGTTTTATAAGGAGCGGGATCCCTTTGACGAATATAATCAGACGTTTGTGATATCGGAAGCCGCTCACTGGGACAGCCAGCAGCAGTTCAAGAAGACTAAGCCGATAGCCATTAAGGAGACTGATGTGGGCGAAGGCTGGGTGAGAGTAGAACTCGCCTGGGCGGTCAGAGTTTGGCTAGAAGGACGCCAAAGATTGCACACATTACACGTCGCGTGCAGGACTTGTCAGCTACGTCGCGCTCCGCTCTCCTTCCATGAGAAGCATCGTCCTTTTCTTGTGCTGTATACCAAATACGCGGGCAAGCGTCGTCGAGGGAGAGCCTTGGAGTGTGATCCTAACACCAACGAGTGTTGTCGCGAGCCCCTATACATCAGCTTCAAGGACCTGGGCTGGGATTACTGGATCATCAGACCGGCTGGATACCACGCGTATTTCTGCAAAGGAAGCTGCGCCCCTATATCTGCGGTCACAACTTCCAAGAGCTACCATCACGACATCATCAGGAAATACTTCTACACTGTGTCGGAACGAGTGGCGGAGTTCAAGCCTTGCTGTGCCCCGACGACCTTCAGCTCCTTACAGCTACTTTACATAGACTCGACGAATACAGTAACGCAAAAGACCCTGCCTAATATGGTGGTCGAATCGTGTGGTTGCATGTGA
Protein
MIAWSTSALVFILVGNFLSLDGEMLGSCANCANADLSVTQLTEARIEFVKQQILKKLRMTKKPAVALPVNSLPMPVAQGQTLSEGTDKPPEFDDYYGRTEQKIIFPVEGECLTSGRYPFKCLQFELPPDVEPEEVSVVELWFYKERDPFDEYNQTFVISEAAHWDSQQQFKKTKPIAIKETDVGEGWVRVELAWAVRVWLEGRQRLHTLHVACRTCQLRRAPLSFHEKHRPFLVLYTKYAGKRRRGRALECDPNTNECCREPLYISFKDLGWDYWIIRPAGYHAYFCKGSCAPISAVTTSKSYHHDIIRKYFYTVSERVAEFKPCCAPTTFSSLQLLYIDSTNTVTQKTLPNMVVESCGCM
Summary
Similarity
Belongs to the TGF-beta family.
Uniprot
H9J528
A0A023GZ29
A0A220QT43
A0A2W1BJ23
A0A0N1IPS4
A0A194QPY5
+ More
A0A0L7KYI2 A0A3S2LVJ7 A0A2H1W2J0 A0A067QL23 A0A2J7QMW1 A0A1Y1KRF1 A0A026W4I5 A0A195DCD0 A0A154PM44 E2C3B4 A0A088AUW0 A0A0L7QL85 D7EL53 A0A2A3E1E4 A0A195BCZ1 A0A195F2T5 A0A0T6ATI5 A0A151WF99 A0A0C9QQQ3 E2AI45 N6T9B8 U4UN74 A0A1W4WRH3 A0A336MR58 A0A336K1S4 A0A1L8DJE8 A0A1L8DJ37 A0A1L8DJ36 A0A1L8DJB8 A0A1J1IDB4 A0A2P8YUF9 F4WM88 A0A1B6DX80 B4HBY4 A0A3B0JPV6 A0A0N0U562 A0A0J7NF23 B4N015 B3ML96 B4KKY5 A0A0M5J422 B4JQ68 E0VAB4 B4M9J2 A0A0L0CMK4 A0A0A1X9X5 B4Q8K8 B4I2Q6 W8AHT8 Q9VQG9 A0A1A9X484 B4NX84 B3NA64 D3TM35 A0A1W4UU43 A0A0K8TX25 A0A034VJT4 B7Q0S9 A0A1Z5KUJ4 A0A2R5LGI8 A0A1I8PKJ1 A0A1I8MMF6 V5IE09 D1LWV4 A0A146LIR5 A0A0K8SRY5 A0A0A9Y4J8 A0A1S3HU52 A0A023G4Y9 A0A0K8VZY0 A0A1D1VAX2 A0A226D1G1 A0A1D1V2H3 A0A210R622 Q0Q617 A0A1W7RA62 U4UA25 Q4SES4 A0A087U896 H3CY16 D2J5T0 A0A3Q3NAP7 A0A3B3DRG6 A0A2D0R5G3 A0A3B5B0P7 A0A1W4XJU5
A0A0L7KYI2 A0A3S2LVJ7 A0A2H1W2J0 A0A067QL23 A0A2J7QMW1 A0A1Y1KRF1 A0A026W4I5 A0A195DCD0 A0A154PM44 E2C3B4 A0A088AUW0 A0A0L7QL85 D7EL53 A0A2A3E1E4 A0A195BCZ1 A0A195F2T5 A0A0T6ATI5 A0A151WF99 A0A0C9QQQ3 E2AI45 N6T9B8 U4UN74 A0A1W4WRH3 A0A336MR58 A0A336K1S4 A0A1L8DJE8 A0A1L8DJ37 A0A1L8DJ36 A0A1L8DJB8 A0A1J1IDB4 A0A2P8YUF9 F4WM88 A0A1B6DX80 B4HBY4 A0A3B0JPV6 A0A0N0U562 A0A0J7NF23 B4N015 B3ML96 B4KKY5 A0A0M5J422 B4JQ68 E0VAB4 B4M9J2 A0A0L0CMK4 A0A0A1X9X5 B4Q8K8 B4I2Q6 W8AHT8 Q9VQG9 A0A1A9X484 B4NX84 B3NA64 D3TM35 A0A1W4UU43 A0A0K8TX25 A0A034VJT4 B7Q0S9 A0A1Z5KUJ4 A0A2R5LGI8 A0A1I8PKJ1 A0A1I8MMF6 V5IE09 D1LWV4 A0A146LIR5 A0A0K8SRY5 A0A0A9Y4J8 A0A1S3HU52 A0A023G4Y9 A0A0K8VZY0 A0A1D1VAX2 A0A226D1G1 A0A1D1V2H3 A0A210R622 Q0Q617 A0A1W7RA62 U4UA25 Q4SES4 A0A087U896 H3CY16 D2J5T0 A0A3Q3NAP7 A0A3B3DRG6 A0A2D0R5G3 A0A3B5B0P7 A0A1W4XJU5
Pubmed
19121390
28756777
26354079
26227816
24845553
28004739
+ More
24508170 30249741 20798317 18362917 19820115 23537049 29403074 21719571 17994087 18057021 20566863 26108605 25830018 22936249 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20353571 25348373 28528879 25315136 25765539 26823975 25401762 27649274 28812685 16837574 15496914 20040489 29451363
24508170 30249741 20798317 18362917 19820115 23537049 29403074 21719571 17994087 18057021 20566863 26108605 25830018 22936249 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20353571 25348373 28528879 25315136 25765539 26823975 25401762 27649274 28812685 16837574 15496914 20040489 29451363
EMBL
BABH01038858
BABH01038859
BABH01038860
BABH01038861
KP861419
AKG96796.1
+ More
JQ768348 AFV61648.1 KY328716 ASK12084.1 KZ150011 PZC75072.1 KQ460077 KPJ17894.1 KQ458671 KPJ05566.1 JTDY01004474 KOB68104.1 RSAL01000181 RVE44930.1 ODYU01005901 SOQ47268.1 KK853224 KDR09701.1 NEVH01013202 PNF29919.1 GEZM01080869 JAV62016.1 KK107488 QOIP01000012 EZA49954.1 RLU15677.1 KQ980989 KYN10543.1 KQ434954 KZC12524.1 GL452318 EFN77560.1 KQ414934 KOC59355.1 DS497806 EFA11884.1 KZ288455 PBC25520.1 KQ976522 KYM82064.1 KQ981855 KYN34773.1 LJIG01022830 KRT78496.1 KQ983219 KYQ46520.1 GBYB01006049 JAG75816.1 GL439639 EFN66896.1 APGK01047280 KB741077 ENN74323.1 KB632432 ERL95539.1 UFQT01002058 SSX32560.1 UFQS01000002 UFQT01000002 SSW96703.1 SSX17090.1 GFDF01007620 JAV06464.1 GFDF01007618 JAV06466.1 GFDF01007621 JAV06463.1 GFDF01007619 JAV06465.1 CVRI01000043 CRK96433.1 PYGN01000354 PSN47866.1 GL888217 EGI64732.1 GEDC01007059 JAS30239.1 CH479272 EDW39995.1 OUUW01000006 SPP82422.1 KQ435794 KOX74121.1 LBMM01005899 KMQ91110.1 CH963920 EDW77950.1 CH902620 EDV30685.1 KPU72990.1 CH933807 EDW11715.1 CP012523 ALC39818.1 CH916372 EDV99048.1 DS235005 EEB10320.1 CH940654 EDW57868.1 KRF77991.1 JRES01000175 KNC33583.1 GBXI01006188 GBXI01006059 GBXI01001358 JAD08104.1 JAD08233.1 JAD12934.1 CM000361 CM002910 EDX03525.1 KMY87746.1 CH480820 EDW54051.1 GAMC01018510 JAB88045.1 AE014134 AY051485 AAF51204.1 AAK92909.1 AAN10416.1 AGB92504.1 CM000157 EDW87441.1 KRJ97158.1 CH954177 EDV57527.1 CCAG010014202 EZ422487 ADD18763.1 GDHF01033280 GDHF01005965 JAI19034.1 JAI46349.1 GAKP01015391 JAC43561.1 ABJB010272678 ABJB010482574 ABJB010734689 ABJB010979147 DS833764 EEC12451.1 GFJQ02008416 JAV98553.1 GGLE01004311 MBY08437.1 GANP01011007 JAB73461.1 GU075931 ACY92460.1 GDHC01011892 GDHC01003268 JAQ06737.1 JAQ15361.1 GBRD01009761 JAG56063.1 GBHO01019144 JAG24460.1 GBBM01006596 JAC28822.1 GDHF01007877 JAI44437.1 BDGG01000003 GAU96013.1 LNIX01000042 OXA39013.1 GAU96014.1 NEDP02000216 OWF56341.1 DQ517928 ABF61781.1 GFAH01000365 JAV48024.1 KB631865 ERL86800.1 CAAE01014614 CAG00858.1 KK118688 KFM73585.1 GQ896540 ACZ60068.1
JQ768348 AFV61648.1 KY328716 ASK12084.1 KZ150011 PZC75072.1 KQ460077 KPJ17894.1 KQ458671 KPJ05566.1 JTDY01004474 KOB68104.1 RSAL01000181 RVE44930.1 ODYU01005901 SOQ47268.1 KK853224 KDR09701.1 NEVH01013202 PNF29919.1 GEZM01080869 JAV62016.1 KK107488 QOIP01000012 EZA49954.1 RLU15677.1 KQ980989 KYN10543.1 KQ434954 KZC12524.1 GL452318 EFN77560.1 KQ414934 KOC59355.1 DS497806 EFA11884.1 KZ288455 PBC25520.1 KQ976522 KYM82064.1 KQ981855 KYN34773.1 LJIG01022830 KRT78496.1 KQ983219 KYQ46520.1 GBYB01006049 JAG75816.1 GL439639 EFN66896.1 APGK01047280 KB741077 ENN74323.1 KB632432 ERL95539.1 UFQT01002058 SSX32560.1 UFQS01000002 UFQT01000002 SSW96703.1 SSX17090.1 GFDF01007620 JAV06464.1 GFDF01007618 JAV06466.1 GFDF01007621 JAV06463.1 GFDF01007619 JAV06465.1 CVRI01000043 CRK96433.1 PYGN01000354 PSN47866.1 GL888217 EGI64732.1 GEDC01007059 JAS30239.1 CH479272 EDW39995.1 OUUW01000006 SPP82422.1 KQ435794 KOX74121.1 LBMM01005899 KMQ91110.1 CH963920 EDW77950.1 CH902620 EDV30685.1 KPU72990.1 CH933807 EDW11715.1 CP012523 ALC39818.1 CH916372 EDV99048.1 DS235005 EEB10320.1 CH940654 EDW57868.1 KRF77991.1 JRES01000175 KNC33583.1 GBXI01006188 GBXI01006059 GBXI01001358 JAD08104.1 JAD08233.1 JAD12934.1 CM000361 CM002910 EDX03525.1 KMY87746.1 CH480820 EDW54051.1 GAMC01018510 JAB88045.1 AE014134 AY051485 AAF51204.1 AAK92909.1 AAN10416.1 AGB92504.1 CM000157 EDW87441.1 KRJ97158.1 CH954177 EDV57527.1 CCAG010014202 EZ422487 ADD18763.1 GDHF01033280 GDHF01005965 JAI19034.1 JAI46349.1 GAKP01015391 JAC43561.1 ABJB010272678 ABJB010482574 ABJB010734689 ABJB010979147 DS833764 EEC12451.1 GFJQ02008416 JAV98553.1 GGLE01004311 MBY08437.1 GANP01011007 JAB73461.1 GU075931 ACY92460.1 GDHC01011892 GDHC01003268 JAQ06737.1 JAQ15361.1 GBRD01009761 JAG56063.1 GBHO01019144 JAG24460.1 GBBM01006596 JAC28822.1 GDHF01007877 JAI44437.1 BDGG01000003 GAU96013.1 LNIX01000042 OXA39013.1 GAU96014.1 NEDP02000216 OWF56341.1 DQ517928 ABF61781.1 GFAH01000365 JAV48024.1 KB631865 ERL86800.1 CAAE01014614 CAG00858.1 KK118688 KFM73585.1 GQ896540 ACZ60068.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000037510
UP000283053
UP000027135
+ More
UP000235965 UP000053097 UP000279307 UP000078492 UP000076502 UP000008237 UP000005203 UP000053825 UP000007266 UP000242457 UP000078540 UP000078541 UP000075809 UP000000311 UP000019118 UP000030742 UP000192223 UP000183832 UP000245037 UP000007755 UP000008744 UP000268350 UP000053105 UP000036403 UP000007798 UP000007801 UP000009192 UP000092553 UP000001070 UP000009046 UP000008792 UP000037069 UP000000304 UP000001292 UP000000803 UP000091820 UP000002282 UP000008711 UP000092444 UP000192221 UP000001555 UP000095300 UP000095301 UP000085678 UP000186922 UP000198287 UP000242188 UP000054359 UP000007303 UP000261640 UP000261560 UP000221080 UP000261400
UP000235965 UP000053097 UP000279307 UP000078492 UP000076502 UP000008237 UP000005203 UP000053825 UP000007266 UP000242457 UP000078540 UP000078541 UP000075809 UP000000311 UP000019118 UP000030742 UP000192223 UP000183832 UP000245037 UP000007755 UP000008744 UP000268350 UP000053105 UP000036403 UP000007798 UP000007801 UP000009192 UP000092553 UP000001070 UP000009046 UP000008792 UP000037069 UP000000304 UP000001292 UP000000803 UP000091820 UP000002282 UP000008711 UP000092444 UP000192221 UP000001555 UP000095300 UP000095301 UP000085678 UP000186922 UP000198287 UP000242188 UP000054359 UP000007303 UP000261640 UP000261560 UP000221080 UP000261400
Interpro
Gene 3D
ProteinModelPortal
H9J528
A0A023GZ29
A0A220QT43
A0A2W1BJ23
A0A0N1IPS4
A0A194QPY5
+ More
A0A0L7KYI2 A0A3S2LVJ7 A0A2H1W2J0 A0A067QL23 A0A2J7QMW1 A0A1Y1KRF1 A0A026W4I5 A0A195DCD0 A0A154PM44 E2C3B4 A0A088AUW0 A0A0L7QL85 D7EL53 A0A2A3E1E4 A0A195BCZ1 A0A195F2T5 A0A0T6ATI5 A0A151WF99 A0A0C9QQQ3 E2AI45 N6T9B8 U4UN74 A0A1W4WRH3 A0A336MR58 A0A336K1S4 A0A1L8DJE8 A0A1L8DJ37 A0A1L8DJ36 A0A1L8DJB8 A0A1J1IDB4 A0A2P8YUF9 F4WM88 A0A1B6DX80 B4HBY4 A0A3B0JPV6 A0A0N0U562 A0A0J7NF23 B4N015 B3ML96 B4KKY5 A0A0M5J422 B4JQ68 E0VAB4 B4M9J2 A0A0L0CMK4 A0A0A1X9X5 B4Q8K8 B4I2Q6 W8AHT8 Q9VQG9 A0A1A9X484 B4NX84 B3NA64 D3TM35 A0A1W4UU43 A0A0K8TX25 A0A034VJT4 B7Q0S9 A0A1Z5KUJ4 A0A2R5LGI8 A0A1I8PKJ1 A0A1I8MMF6 V5IE09 D1LWV4 A0A146LIR5 A0A0K8SRY5 A0A0A9Y4J8 A0A1S3HU52 A0A023G4Y9 A0A0K8VZY0 A0A1D1VAX2 A0A226D1G1 A0A1D1V2H3 A0A210R622 Q0Q617 A0A1W7RA62 U4UA25 Q4SES4 A0A087U896 H3CY16 D2J5T0 A0A3Q3NAP7 A0A3B3DRG6 A0A2D0R5G3 A0A3B5B0P7 A0A1W4XJU5
A0A0L7KYI2 A0A3S2LVJ7 A0A2H1W2J0 A0A067QL23 A0A2J7QMW1 A0A1Y1KRF1 A0A026W4I5 A0A195DCD0 A0A154PM44 E2C3B4 A0A088AUW0 A0A0L7QL85 D7EL53 A0A2A3E1E4 A0A195BCZ1 A0A195F2T5 A0A0T6ATI5 A0A151WF99 A0A0C9QQQ3 E2AI45 N6T9B8 U4UN74 A0A1W4WRH3 A0A336MR58 A0A336K1S4 A0A1L8DJE8 A0A1L8DJ37 A0A1L8DJ36 A0A1L8DJB8 A0A1J1IDB4 A0A2P8YUF9 F4WM88 A0A1B6DX80 B4HBY4 A0A3B0JPV6 A0A0N0U562 A0A0J7NF23 B4N015 B3ML96 B4KKY5 A0A0M5J422 B4JQ68 E0VAB4 B4M9J2 A0A0L0CMK4 A0A0A1X9X5 B4Q8K8 B4I2Q6 W8AHT8 Q9VQG9 A0A1A9X484 B4NX84 B3NA64 D3TM35 A0A1W4UU43 A0A0K8TX25 A0A034VJT4 B7Q0S9 A0A1Z5KUJ4 A0A2R5LGI8 A0A1I8PKJ1 A0A1I8MMF6 V5IE09 D1LWV4 A0A146LIR5 A0A0K8SRY5 A0A0A9Y4J8 A0A1S3HU52 A0A023G4Y9 A0A0K8VZY0 A0A1D1VAX2 A0A226D1G1 A0A1D1V2H3 A0A210R622 Q0Q617 A0A1W7RA62 U4UA25 Q4SES4 A0A087U896 H3CY16 D2J5T0 A0A3Q3NAP7 A0A3B3DRG6 A0A2D0R5G3 A0A3B5B0P7 A0A1W4XJU5
PDB
5HLY
E-value=1.21066e-31,
Score=340
Ontologies
KEGG
GO
GO:0008083
GO:0005576
GO:0016021
GO:0060395
GO:0042981
GO:0005615
GO:0043408
GO:0010862
GO:0005125
GO:0048468
GO:0005160
GO:0008234
GO:0007411
GO:0008340
GO:0032927
GO:0050796
GO:0010507
GO:0032924
GO:0001533
GO:0030216
GO:0005179
GO:0016887
GO:0006810
GO:0042626
GO:0001678
GO:0005536
GO:0046835
GO:0006281
GO:0006259
PANTHER
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 22,
Likelihood: 0.745368
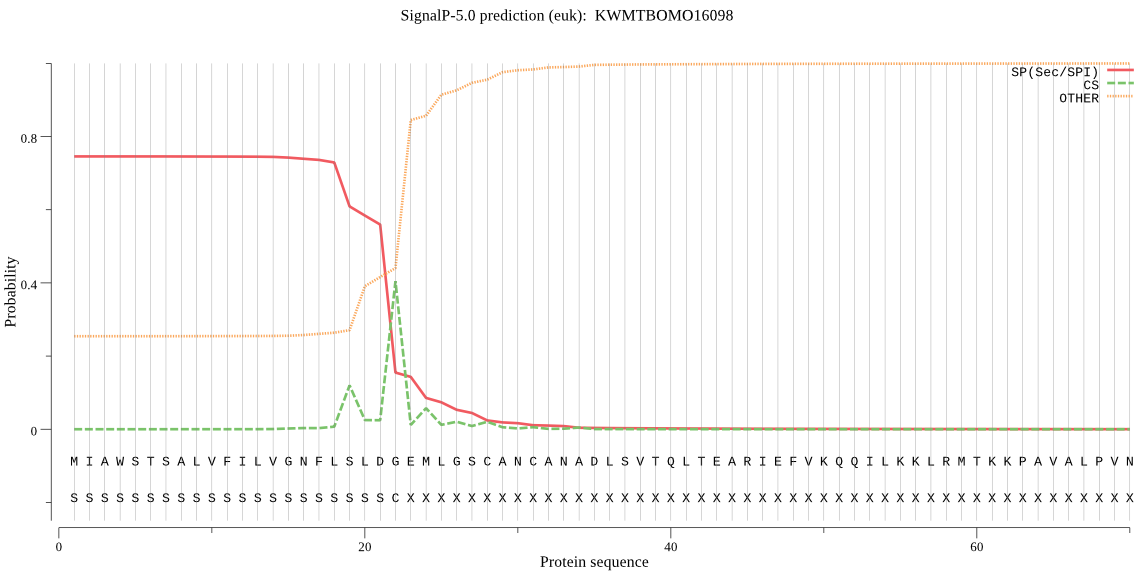
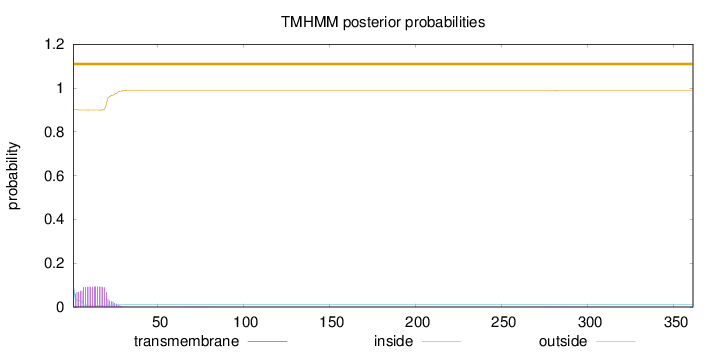
Length:
361
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.78361
Exp number, first 60 AAs:
1.7746
Total prob of N-in:
0.09875
outside
1 - 361
Population Genetic Test Statistics
Pi
217.064572
Theta
162.048898
Tajima's D
1.290298
CLR
0.19951
CSRT
0.733313334333283
Interpretation
Uncertain
