Gene
KWMTBOMO16091 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014536
Annotation
heat_shock_protein_70_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.705
Sequence
CDS
ATGCCAGCTATCGGGATCGATTTGGGAACGACATACTCATGTGTTGGAGTATGGCAGCACGGGAACGTGGAGATCATCGCGAACGACCAGGGCAACCGTACTACACCATCGTACGTCGCGTTCACGGACACGGAGCGTCTCATCGGCGACGCAGCCAAGAACCAGGTCGCCTTGAACCCTAACAACACCGTGTTCGACGCGAAGAGGCTGATCGGGAGGAAATTCGACGACCCCAAGATTCAGCAGGACATGAAGCACTGGCCCTTCAAAGTGATCAACGACTGCGGCAAACCGAAAATACAGATCGAGTTCAAAGGTGAGACGAAACGATTTGCGCCAGAAGAAATTAGCAGCATGGTGCTGACAAAAATGAAGGAGACGGCGGAAGCGTATCTGGGAAGCACAGTGCGGGATGCGGTAGTCACAGTTCCGGCATACTTCAACGACTCCCAGCGTCAGGCCACCAAGGACGCCGGAGCCATCGCCGGCCTGAACGTGCTCCGCATCATCAACGAGCCCACAGCCGCCGCGCTCGCGTACGGCCTCGACAAGAACCTCAAGGGTGAGAGAAATGTTCTTATCTTCGATCTGGGTGGAGGAACCTTCGACGTGTCCGTCCTGACCATCGACGAGGGATCGTTGTTCGAAGTGAAGTCCACAGCCGGCGACACGCACCTGGGAGGCGAGGACTTCGATAACCGACTAGTCAACCATCTTGCGGAAGAGTTCAAGCGCAAGTACAAGAAGGATCTGCGCCTGAACTCTCGCGCACTCCGACGCCTCCGCACGGCCGCTGAGCGCGCCAAGAGGACACTCTCATCCAGCACCGAGGCCACCATCGAGATTGACGCTCTGTATGAGGGCATCGACTTCTACACGCGAGTCTCCCGCGCCCGCTTCGAGGAACTGAACGCGGACCTGTTCAGGGGAACTCTGGAACCCGTCGAGAAGGCACTCAAGGATGCTAAACTCGACAAGAGTCAGATCCACGACGTGGTCCTCGTCGGAGGCTCCACCCGCATTCCGAAAGTACAGACTATGCTCCAAAACTTCTTCTGTGGGAAGAAACTGAACCTATCCATCAATCCGGACGAAGCCGTAGCCTACGGTGCAGCAGTCCAGGCGGCCATCCTGAGCGGCGAGACCGACTCCAAGATACAGGACGTCTTGTTGGTCGACGTGGCTCCTCTGTCTCTCGGTATTGAGACAGCCGGCGGCGTGATGACGAAGATCATCGAACGAAACTCAAAGATTCCGTGCAAACAGTCTCAGACATTCACGACGTACTCAGACAACCAGCCGGCCGTCACCATCCAGGTGTACGAGGGAGAGAGAGCGATGACAAAGGACAACAACCTTCTGGGCACGTTCGACCTGACGGGGATCCCGCCCGCGCCGCGAGGAGTGCCCAAGATCGACGTGACGTTCGACATGGACGCTAACGGCATCCTGAACGTGTCGGCCAAGGAGAACAGCACCGGCCGCAGCAAGAACATCGTGATCAAGAACGACAAGGGTCGTCTCTCGCAAGCGGAGATCGATCGCATGCTGTCCGAGGCAGAGCGGTACAAGGAGGAAGACGAGAAGCAGAGACAGCGCGTGGCCGCTCGGAACCAGCTCGAATCGTATTTGTTCAGCGTGAAGCAGGCGCTCGACGAGGCCGGCGACAAACTGAGCGACGCGGACAAGAGCACGGCGCGCGACGCGTGTGACGAGGCGCTGCGGTGGCTGGACAACAACACTCTCGCTGACCAGGATGAATACGAGCACAAGCTGAAGGATGTGCAGCGAGTGTGTTCGCCGGTCATGAGCAAGATGCACGGTGCGGCACCCGGCGGTATGCCAGGAGGAATGCCGGGTGGCATGCCAGGAGGAATGTCAGGAGGTATGCCAGGAGGCATGCCAGGAGGAATGCCGGGTGGCATGCCGGGAGGTATGCCGGGCGGTATGCCCGGATACATGCCTGGTGGAATGCCGGGTGGATACCAAATGCCAAGAAATACTGGACCCACCGTTGAAGAAGTCGATTGA
Protein
MPAIGIDLGTTYSCVGVWQHGNVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVALNPNNTVFDAKRLIGRKFDDPKIQQDMKHWPFKVINDCGKPKIQIEFKGETKRFAPEEISSMVLTKMKETAEAYLGSTVRDAVVTVPAYFNDSQRQATKDAGAIAGLNVLRIINEPTAAALAYGLDKNLKGERNVLIFDLGGGTFDVSVLTIDEGSLFEVKSTAGDTHLGGEDFDNRLVNHLAEEFKRKYKKDLRLNSRALRRLRTAAERAKRTLSSSTEATIEIDALYEGIDFYTRVSRARFEELNADLFRGTLEPVEKALKDAKLDKSQIHDVVLVGGSTRIPKVQTMLQNFFCGKKLNLSINPDEAVAYGAAVQAAILSGETDSKIQDVLLVDVAPLSLGIETAGGVMTKIIERNSKIPCKQSQTFTTYSDNQPAVTIQVYEGERAMTKDNNLLGTFDLTGIPPAPRGVPKIDVTFDMDANGILNVSAKENSTGRSKNIVIKNDKGRLSQAEIDRMLSEAERYKEEDEKQRQRVAARNQLESYLFSVKQALDEAGDKLSDADKSTARDACDEALRWLDNNTLADQDEYEHKLKDVQRVCSPVMSKMHGAAPGGMPGGMPGGMPGGMSGGMPGGMPGGMPGGMPGGMPGGMPGYMPGGMPGGYQMPRNTGPTVEEVD
Summary
Similarity
Belongs to the heat shock protein 70 family.
Uniprot
Q2F683
F8UN42
A6P319
C4P3P9
C0LRA8
A0A2C9PHS1
+ More
E7D2J5 Q6L6T3 A0A212F995 T1SC34 T1SBT7 A0A2H1W9K6 T1SBT5 A0A194QJ85 D9D840 T1SBT1 C8CCR3 E3UVB8 A0A212FCV0 E0A3F8 J7IL09 E0A3F7 A0A0L7KZH8 E0A3F6 A0A2W1BCN7 Q0KKB4 A0A0S1MMB1 U3KZM6 C6KEP2 B1A8R7 J7IIA2 E9LK30 E9LK31 F8UN43 J7I5M9 A9CSM0 A5A3D9 Q86GI6 I3WA85 I3WA86 E2FZ92 A0A068F6C3 E2GLM4 J7FIB4 D1CSA1 D6QX79 B5TGR3 J7G295 D1LWU2 B5AN02 B5TGQ9 A0A023F633 T1I0D9 A0A1Z1JMY7 A0A023F628 J3JWS1 A5HTV3 A0A1B6KRC2 A0A172JCK2 A0A1S5VZD3 A0A1L2EC59 A0A346THN6 B4K7C9 A0A224XGB7 A0A346THM2 F5BYS7 A0A165BK16 A0A1S3D948 D6WU59 W5RXA8 D6WT72 J9JMZ6 A0A3G1RKN6 K9MV11 A0A1Z1JNE2 F1C382 A0A1I8P0G5 A0A1S3DAN2 A0A1Z1JMY1 A0A2P0XIF6 A0A0B4L040 A0A1S3D964 N6TG46 F1C378 U3GPH7 A0A1B6DH77 A0A0N1IFZ8 F1C379 A0A212F282 A0A1S5VZG2 F1C380 Q4ZGJ2 A0A172JCK1 A0A240FEJ6 Q5I938 A0A1W4XW28 A0A2J7PG88 D6QX82 D9IFR9 A0A067QTK9
E7D2J5 Q6L6T3 A0A212F995 T1SC34 T1SBT7 A0A2H1W9K6 T1SBT5 A0A194QJ85 D9D840 T1SBT1 C8CCR3 E3UVB8 A0A212FCV0 E0A3F8 J7IL09 E0A3F7 A0A0L7KZH8 E0A3F6 A0A2W1BCN7 Q0KKB4 A0A0S1MMB1 U3KZM6 C6KEP2 B1A8R7 J7IIA2 E9LK30 E9LK31 F8UN43 J7I5M9 A9CSM0 A5A3D9 Q86GI6 I3WA85 I3WA86 E2FZ92 A0A068F6C3 E2GLM4 J7FIB4 D1CSA1 D6QX79 B5TGR3 J7G295 D1LWU2 B5AN02 B5TGQ9 A0A023F633 T1I0D9 A0A1Z1JMY7 A0A023F628 J3JWS1 A5HTV3 A0A1B6KRC2 A0A172JCK2 A0A1S5VZD3 A0A1L2EC59 A0A346THN6 B4K7C9 A0A224XGB7 A0A346THM2 F5BYS7 A0A165BK16 A0A1S3D948 D6WU59 W5RXA8 D6WT72 J9JMZ6 A0A3G1RKN6 K9MV11 A0A1Z1JNE2 F1C382 A0A1I8P0G5 A0A1S3DAN2 A0A1Z1JMY1 A0A2P0XIF6 A0A0B4L040 A0A1S3D964 N6TG46 F1C378 U3GPH7 A0A1B6DH77 A0A0N1IFZ8 F1C379 A0A212F282 A0A1S5VZG2 F1C380 Q4ZGJ2 A0A172JCK1 A0A240FEJ6 Q5I938 A0A1W4XW28 A0A2J7PG88 D6QX82 D9IFR9 A0A067QTK9
Pubmed
EMBL
DQ311189
ABD36134.1
JF836794
AEI58996.1
AB035326
BAF69068.1
+ More
FJ862049 ACQ78180.1 FJ754276 ACN78407.1 KY930331 ATL76715.1 HQ012004 ADV03160.1 AB179657 BAD18974.1 AGBW02009614 OWR50325.1 JX680669 AGR84216.1 JX680670 AGR84217.1 ODYU01007205 SOQ49779.1 JX680677 AGR84224.1 KQ458671 KPJ05577.1 GU945198 KR821069 ADI50267.1 ALL42057.1 JX680676 AGR84223.1 GQ389711 ACV32640.1 HM593518 ADP37711.1 AGBW02009136 OWR51575.1 HM370511 ADK94699.1 JQ693014 AFQ37587.1 HM370510 ADK94698.1 JTDY01004123 KOB68565.1 HM370509 ADK94697.1 KZ150133 PZC73022.1 AB251895 BAF03555.1 KT225460 ALL42052.1 JX678981 AFU06382.1 GQ199477 ACS72236.1 EU430480 ABZ10939.1 JQ693015 AFQ37588.1 HQ107971 ADV58254.1 HQ107972 ADV58255.1 BABH01042878 JF836795 AEI58997.1 JQ711194 AFQ33498.1 AB325801 BAF95560.1 EF523381 ABP93405.1 AY220911 AAO65964.1 JN863694 AFK93489.2 JN863695 AFK93490.1 HM212645 ADK39311.1 KJ701421 AID61521.1 HM367079 ADO14473.1 JQ316541 AFO70209.1 DQ093385 ACZ52196.1 HM013709 ADG03465.1 EU934244 ACH85201.1 JX112893 AFP54305.1 GU074513 ACY71070.1 EU861391 ACF74975.2 EU934240 ACH85197.1 GBBI01002049 JAC16663.1 ACPB03005879 KY705385 ARW29611.1 GBBI01002050 JAC16662.1 APGK01039300 BT127689 KB740969 KB632230 AEE62651.1 ENN76739.1 ERL90378.1 EF569673 ABQ39970.1 GEBQ01026193 JAT13784.1 KU311038 AMQ77288.1 KX695168 AQP31338.1 KT003964 ANJ86404.1 MG265913 AXU24969.1 CH933806 EDW14253.1 GFTR01007568 JAW08858.1 MG265899 AXU24955.1 JF421286 AEB52075.1 KU159184 AMX23517.1 KQ971352 EFA07459.1 EFA07510.1 KF771049 AHG94986.1 KQ971354 EFA07173.1 ABLF02040542 MF773976 AXB26576.1 JX627810 AFX84560.1 KY705383 ARW29609.1 HQ184405 ADX42270.1 KY705384 ARW29610.1 KY661325 AVA17412.1 KC620440 AHE77387.1 APGK01039299 ENN76738.1 ERL90377.1 HQ184404 ADX42267.1 KF018929 AGT99186.1 GEDC01012328 JAS24970.1 KQ460126 KPJ17595.1 ADX42268.1 AGBW02010783 OWR47855.1 KX976473 AQP31363.1 ADX42269.1 DQ017057 AAY28732.1 KU311037 AMQ77287.1 KY231148 ARQ84028.1 AY842477 AAW32099.2 NEVH01025635 PNF15343.1 HM013712 ADG03468.1 HM141537 ADJ96611.1 KK853289 KDR08926.1
FJ862049 ACQ78180.1 FJ754276 ACN78407.1 KY930331 ATL76715.1 HQ012004 ADV03160.1 AB179657 BAD18974.1 AGBW02009614 OWR50325.1 JX680669 AGR84216.1 JX680670 AGR84217.1 ODYU01007205 SOQ49779.1 JX680677 AGR84224.1 KQ458671 KPJ05577.1 GU945198 KR821069 ADI50267.1 ALL42057.1 JX680676 AGR84223.1 GQ389711 ACV32640.1 HM593518 ADP37711.1 AGBW02009136 OWR51575.1 HM370511 ADK94699.1 JQ693014 AFQ37587.1 HM370510 ADK94698.1 JTDY01004123 KOB68565.1 HM370509 ADK94697.1 KZ150133 PZC73022.1 AB251895 BAF03555.1 KT225460 ALL42052.1 JX678981 AFU06382.1 GQ199477 ACS72236.1 EU430480 ABZ10939.1 JQ693015 AFQ37588.1 HQ107971 ADV58254.1 HQ107972 ADV58255.1 BABH01042878 JF836795 AEI58997.1 JQ711194 AFQ33498.1 AB325801 BAF95560.1 EF523381 ABP93405.1 AY220911 AAO65964.1 JN863694 AFK93489.2 JN863695 AFK93490.1 HM212645 ADK39311.1 KJ701421 AID61521.1 HM367079 ADO14473.1 JQ316541 AFO70209.1 DQ093385 ACZ52196.1 HM013709 ADG03465.1 EU934244 ACH85201.1 JX112893 AFP54305.1 GU074513 ACY71070.1 EU861391 ACF74975.2 EU934240 ACH85197.1 GBBI01002049 JAC16663.1 ACPB03005879 KY705385 ARW29611.1 GBBI01002050 JAC16662.1 APGK01039300 BT127689 KB740969 KB632230 AEE62651.1 ENN76739.1 ERL90378.1 EF569673 ABQ39970.1 GEBQ01026193 JAT13784.1 KU311038 AMQ77288.1 KX695168 AQP31338.1 KT003964 ANJ86404.1 MG265913 AXU24969.1 CH933806 EDW14253.1 GFTR01007568 JAW08858.1 MG265899 AXU24955.1 JF421286 AEB52075.1 KU159184 AMX23517.1 KQ971352 EFA07459.1 EFA07510.1 KF771049 AHG94986.1 KQ971354 EFA07173.1 ABLF02040542 MF773976 AXB26576.1 JX627810 AFX84560.1 KY705383 ARW29609.1 HQ184405 ADX42270.1 KY705384 ARW29610.1 KY661325 AVA17412.1 KC620440 AHE77387.1 APGK01039299 ENN76738.1 ERL90377.1 HQ184404 ADX42267.1 KF018929 AGT99186.1 GEDC01012328 JAS24970.1 KQ460126 KPJ17595.1 ADX42268.1 AGBW02010783 OWR47855.1 KX976473 AQP31363.1 ADX42269.1 DQ017057 AAY28732.1 KU311037 AMQ77287.1 KY231148 ARQ84028.1 AY842477 AAW32099.2 NEVH01025635 PNF15343.1 HM013712 ADG03468.1 HM141537 ADJ96611.1 KK853289 KDR08926.1
Proteomes
Pfam
PF00012 HSP70
Interpro
Gene 3D
ProteinModelPortal
Q2F683
F8UN42
A6P319
C4P3P9
C0LRA8
A0A2C9PHS1
+ More
E7D2J5 Q6L6T3 A0A212F995 T1SC34 T1SBT7 A0A2H1W9K6 T1SBT5 A0A194QJ85 D9D840 T1SBT1 C8CCR3 E3UVB8 A0A212FCV0 E0A3F8 J7IL09 E0A3F7 A0A0L7KZH8 E0A3F6 A0A2W1BCN7 Q0KKB4 A0A0S1MMB1 U3KZM6 C6KEP2 B1A8R7 J7IIA2 E9LK30 E9LK31 F8UN43 J7I5M9 A9CSM0 A5A3D9 Q86GI6 I3WA85 I3WA86 E2FZ92 A0A068F6C3 E2GLM4 J7FIB4 D1CSA1 D6QX79 B5TGR3 J7G295 D1LWU2 B5AN02 B5TGQ9 A0A023F633 T1I0D9 A0A1Z1JMY7 A0A023F628 J3JWS1 A5HTV3 A0A1B6KRC2 A0A172JCK2 A0A1S5VZD3 A0A1L2EC59 A0A346THN6 B4K7C9 A0A224XGB7 A0A346THM2 F5BYS7 A0A165BK16 A0A1S3D948 D6WU59 W5RXA8 D6WT72 J9JMZ6 A0A3G1RKN6 K9MV11 A0A1Z1JNE2 F1C382 A0A1I8P0G5 A0A1S3DAN2 A0A1Z1JMY1 A0A2P0XIF6 A0A0B4L040 A0A1S3D964 N6TG46 F1C378 U3GPH7 A0A1B6DH77 A0A0N1IFZ8 F1C379 A0A212F282 A0A1S5VZG2 F1C380 Q4ZGJ2 A0A172JCK1 A0A240FEJ6 Q5I938 A0A1W4XW28 A0A2J7PG88 D6QX82 D9IFR9 A0A067QTK9
E7D2J5 Q6L6T3 A0A212F995 T1SC34 T1SBT7 A0A2H1W9K6 T1SBT5 A0A194QJ85 D9D840 T1SBT1 C8CCR3 E3UVB8 A0A212FCV0 E0A3F8 J7IL09 E0A3F7 A0A0L7KZH8 E0A3F6 A0A2W1BCN7 Q0KKB4 A0A0S1MMB1 U3KZM6 C6KEP2 B1A8R7 J7IIA2 E9LK30 E9LK31 F8UN43 J7I5M9 A9CSM0 A5A3D9 Q86GI6 I3WA85 I3WA86 E2FZ92 A0A068F6C3 E2GLM4 J7FIB4 D1CSA1 D6QX79 B5TGR3 J7G295 D1LWU2 B5AN02 B5TGQ9 A0A023F633 T1I0D9 A0A1Z1JMY7 A0A023F628 J3JWS1 A5HTV3 A0A1B6KRC2 A0A172JCK2 A0A1S5VZD3 A0A1L2EC59 A0A346THN6 B4K7C9 A0A224XGB7 A0A346THM2 F5BYS7 A0A165BK16 A0A1S3D948 D6WU59 W5RXA8 D6WT72 J9JMZ6 A0A3G1RKN6 K9MV11 A0A1Z1JNE2 F1C382 A0A1I8P0G5 A0A1S3DAN2 A0A1Z1JMY1 A0A2P0XIF6 A0A0B4L040 A0A1S3D964 N6TG46 F1C378 U3GPH7 A0A1B6DH77 A0A0N1IFZ8 F1C379 A0A212F282 A0A1S5VZG2 F1C380 Q4ZGJ2 A0A172JCK1 A0A240FEJ6 Q5I938 A0A1W4XW28 A0A2J7PG88 D6QX82 D9IFR9 A0A067QTK9
PDB
5FPN
E-value=0,
Score=2209
Ontologies
PATHWAY
GO
PANTHER
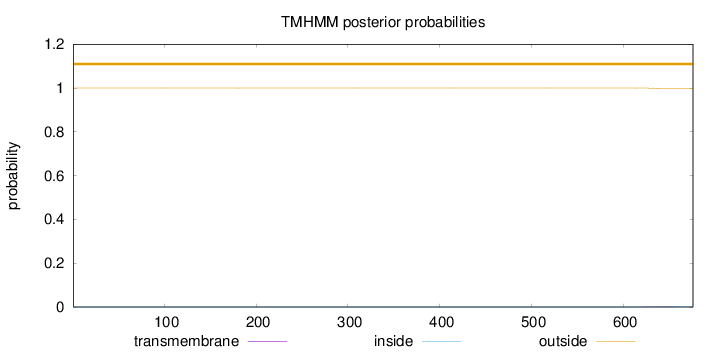
Topology
Length:
676
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04874
Exp number, first 60 AAs:
0.00041
Total prob of N-in:
0.00021
outside
1 - 676
Population Genetic Test Statistics
Pi
464.086553
Theta
217.853018
Tajima's D
3.50943
CLR
0.125546
CSRT
0.994000299985001
Interpretation
Uncertain
