Gene
KWMTBOMO16087 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004612
Annotation
heat_shock_protein_83_[Bombyx_mori]
Full name
Heat shock protein 83
Location in the cell
Cytoplasmic Reliability : 3.856
Sequence
CDS
ATGCCGGAAGAAATGGAGACACAGCCAGCGGAGGTTGAAACCTTCGCGTTCCAGGCTGAGATCGCTCAGCTTATGTCCCTGATCATCAACACCTTTTACTCCAACAAAGAAATTTTCCTTCGTGAGCTGATTTCCAATTCATCGGACGCTTTAGACAAAATCAGGTATGAATCTCTCACGGATCCGTCAAAACTCGATAGTGGCAAAGAGCTGTACATCAAGATCATTCCCAACAAGAACGAGGGCACTCTTACGATCATCGATACCGGTATTGGTATGACCAAGGCCGATTTGGTGAACAATTTGGGAACCATCGCGAAATCTGGTACTAAAGCTTTCATGGAGGCTCTTCAAGCAGGTGCCGACATCAGCATGATTGGACAGTTCGGTGTTGGCTTCTACTCCAGTTACTTGGTCGCTGACCGCGTGACTGTTCACTCTAAACACAATGACGACGAGCAATACGTGTGGGAATCTTCTGCAGGAGGCTCGTTCACAGTCCGCCCAGACAGCGGTGAGCCCCTTGGTCGAGGTACAAAGATCGTCCTTCACGTCAAAGAGGACTTGGCAGAATTCATGGAAGAACACAAAATCAAAGAGATCGTAAAGAAACATTCCCAGTTCATCGGCTACCCAATCAAGCTGATGGTTGAAAAAGAACGCGAAAAAGAACTGTCTGATGATGAAGCCGAAGAAGAAAAGAAGGAAGAGGAAGATGAGAAGCCTAAGATTGAAGATGTAGGTGAGGATGAAGACGAGGACAAAAAAGATACGAAGAAGAAGAAGAAAACAATTAAAGAAAAATACACAGAGGATGAAGAACTCAACAAGACAAAGCCTATCTGGACCAGAAACGCTGATGACATTACCCAAGATGAGTATGGAGACTTCTACAAATCCCTTACCAATGATTGGGAGGACCATCTTGCAGTCAAACACTTCTCCGTTGAAGGTCAGTTAGAGTTCCGTGCACTACTTTTTGTGCCACGTCGTGCACCTTTTGACTTGTTTGAAAACAAGAAGCGCAAGAACAATATCAAGCTGTATGTACGAAGAGTTTTCATCATGGACAACTGTGAGGATCTCATCCCAGAGTACCTTAACTTCATTAGAGGTGTTGTCGACAGTGAGGACCTTCCTCTAAACATTTCACGTGAGATGCTTCAACAGAATAAAATTCTGAAAGTAATTAGGAAGAATTTAGTTAAAAAATGCCTAGAACTATTTGAAGAATTGGCAGAGGACAAAGAAAACTACAAGAAGTATTATGAACAATTCAGCAAGAACTTAAAATTGGGTATCCATGAAGACTCTCAAAACAGGGCTAAGCTCTCTGAGCTTCTGCGGTACCACACTTCGGCATCGGGTGATGAAGCCTGCTCTCTGAAAGAGTATGTATCCAGGATGAAGGAGAATCAGAAACATATCTACTACATCACTGGAGAAAACAGAGACCAGGTGGCGAACTCCTCATTTGTGGAGAGAGTGAAGAAACGTGGCTATGAAGTAGTCTACATGACTGAGCCCATTGATGAATATGTAGTACAACAGATGAGGGAGTATGATGGCAAGACCCTTGTATCGGTCACAAAGGAAGGATTAGAACTTCCTGAAGATGAGGAAGAAAAGAAGAAACGTGAGGAAGATAAGGTGAAGTTCGAAGGGCTGTGCAAGGTGATGAAGAACATCCTGGACAACAAAGTTGAGAAAGTTGTTGTCTCTAACAGACTTGTGGAGTCTCCTTGCTGTATTGTTACTGCTCAATATGGATGGTCTGCCAACATGGAACGTATCATGAAGGCTCAGGCTCTACGTGACACATCCACAATGGGATACATGGCTGCGAAGAAGCATTTGGAAATCAATCCTGATCACTCGATTGTGGAAACATTAAGGCAGAAAGCGGAAGCCGACAAGAATGACAAAGCTGTTAAGGATCTTGTTATCTTATTGTATGAAACTGCCCTGCTGTCTTCTGGCTTCACGCTTGATGAGCCCCAAGTCCATGCTTCCCGTATCTATAGAATGATCAAACTGGGTCTTGGTATTGATGAAGATGAGCCGATTCAGGTTGAAGAGCCTGCTTCTGGAGATGTGCCGCCATTGGAGGGTGATGCCGATGATGCATCTCGCATGGAGGAGGTTGATTAA
Protein
MPEEMETQPAEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIRYESLTDPSKLDSGKELYIKIIPNKNEGTLTIIDTGIGMTKADLVNNLGTIAKSGTKAFMEALQAGADISMIGQFGVGFYSSYLVADRVTVHSKHNDDEQYVWESSAGGSFTVRPDSGEPLGRGTKIVLHVKEDLAEFMEEHKIKEIVKKHSQFIGYPIKLMVEKEREKELSDDEAEEEKKEEEDEKPKIEDVGEDEDEDKKDTKKKKKTIKEKYTEDEELNKTKPIWTRNADDITQDEYGDFYKSLTNDWEDHLAVKHFSVEGQLEFRALLFVPRRAPFDLFENKKRKNNIKLYVRRVFIMDNCEDLIPEYLNFIRGVVDSEDLPLNISREMLQQNKILKVIRKNLVKKCLELFEELAEDKENYKKYYEQFSKNLKLGIHEDSQNRAKLSELLRYHTSASGDEACSLKEYVSRMKENQKHIYYITGENRDQVANSSFVERVKKRGYEVVYMTEPIDEYVVQQMREYDGKTLVSVTKEGLELPEDEEEKKKREEDKVKFEGLCKVMKNILDNKVEKVVVSNRLVESPCCIVTAQYGWSANMERIMKAQALRDTSTMGYMAAKKHLEINPDHSIVETLRQKAEADKNDKAVKDLVILLYETALLSSGFTLDEPQVHASRIYRMIKLGLGIDEDEPIQVEEPASGDVPPLEGDADDASRMEEVD
Summary
Description
Molecular chaperone that promotes the maturation, structural maintenance and proper regulation of specific target proteins involved for instance in cell cycle control and signal transduction. Undergoes a functional cycle that is linked to its ATPase activity. This cycle probably induces conformational changes in the client proteins, thereby causing their activation. Interacts dynamically with various co-chaperones that modulate its substrate recognition, ATPase cycle and chaperone function (By similarity). Required for piRNA biogenesis by facilitating loading of piRNAs into PIWI proteins.
Molecular chaperone that promotes the maturation, structural maintenance and proper regulation of specific target proteins involved for instance in cell cycle control and signal transduction. Undergoes a functional cycle that is linked to its ATPase activity. This cycle probably induces conformational changes in the client proteins, thereby causing their activation. Interacts dynamically with various co-chaperones that modulate its substrate recognition, ATPase cycle and chaperone function. Required for piRNA biogenesis by facilitating loading of piRNAs into PIWI proteins (By similarity).
Molecular chaperone that promotes the maturation, structural maintenance and proper regulation of specific target proteins involved for instance in cell cycle control and signal transduction. Undergoes a functional cycle that is linked to its ATPase activity. This cycle probably induces conformational changes in the client proteins, thereby causing their activation. Interacts dynamically with various co-chaperones that modulate its substrate recognition, ATPase cycle and chaperone function. Required for piRNA biogenesis by facilitating loading of piRNAs into PIWI proteins (By similarity).
Subunit
Homodimer.
Similarity
Belongs to the heat shock protein 90 family.
Keywords
ATP-binding
Chaperone
Complete proteome
Cytoplasm
Nucleotide-binding
Phosphoprotein
Reference proteome
Stress response
Feature
chain Heat shock protein 83
Uniprot
EMBL
AB060275
GU324473
HQ437671
GU230736
ADM26739.1
KU522474
+ More
APX61060.1 GU235994 ADD91573.1 EF523380 ABP93404.1 KJ679917 AJK31515.1 AB176669 BAD15163.1 MG520364 AWX67685.1 AEB39782.1 KC620427 AHE77376.1 CH478474 CH477789 EAT32919.1 EAT36187.1 KX358700 AOQ25849.1 EAT36186.1 AAAB01008807 GDIQ01096087 JAL55639.1 GDIQ01216643 JAK35082.1
APX61060.1 GU235994 ADD91573.1 EF523380 ABP93404.1 KJ679917 AJK31515.1 AB176669 BAD15163.1 MG520364 AWX67685.1 AEB39782.1 KC620427 AHE77376.1 CH478474 CH477789 EAT32919.1 EAT36187.1 KX358700 AOQ25849.1 EAT36186.1 AAAB01008807 GDIQ01096087 JAL55639.1 GDIQ01216643 JAK35082.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
PDB
2CG9
E-value=0,
Score=1817
Ontologies
KEGG
GO
PANTHER
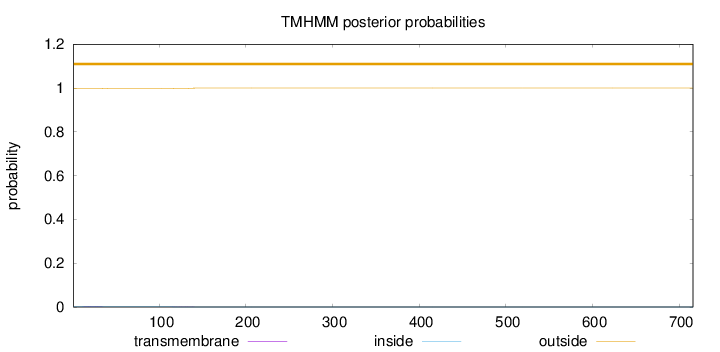
Topology
Subcellular location
Cytoplasm
Length:
716
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0873800000000001
Exp number, first 60 AAs:
0.05878
Total prob of N-in:
0.00316
outside
1 - 716
Population Genetic Test Statistics
Pi
228.777162
Theta
182.790112
Tajima's D
0.21404
CLR
0.345597
CSRT
0.429678516074196
Interpretation
Uncertain
