Gene
KWMTBOMO16082
Annotation
PREDICTED:_transcription_factor_EB-like_[Bombyx_mori]
Full name
Alkaline phosphatase
Location in the cell
Nuclear Reliability : 2.184
Sequence
CDS
ATGACCAAGGGCCCTCGGAAGGTGAAGCTAGTCATTGTAGTCAACAATAAGAAAGACCCGCCTACGTTTAAGACCCTGACGCCCACATCTCGCACACAACTGAAGCAGCAGTTGATGCGAGAACATGCCCAGGAGCAGCTGCGAAGAGAATCACAAAACCAACAAGCTGGACAATCCAAAGACAATGAAGAGCATAAAAAGAAGTCCTCGCCGACGGAGGTGCCTAGGATTACGCCACACGTCGAATTACCGCCACAAGTCCTACAGGTCCGAACAGTTCTGGAGAACCCCACCAGGTATCACGTGATACAGAAGCAGAAGAGTCAGGTACGACAGTACCTCAGCGAGTCCTTCCAACCACACACACAGGTCAGTGAAGTTTGGTCGTTTTTTTAA
Protein
MTKGPRKVKLVIVVNNKKDPPTFKTLTPTSRTQLKQQLMREHAQEQLRRESQNQQAGQSKDNEEHKKKSSPTEVPRITPHVELPPQVLQVRTVLENPTRYHVIQKQKSQVRQYLSESFQPHTQVSEVWSFF
Summary
Catalytic Activity
a phosphate monoester + H2O = an alcohol + phosphate
Similarity
Belongs to the alkaline phosphatase family.
Uniprot
A0A2A4KA52
A0A2W1BFJ0
A0A3S2TFY2
A0A194QJM4
A0A0N0PFG3
A0A212FK81
+ More
A0A088AUU6 A0A2A3E1E0 A0A154PKW9 A0A0C9R0A6 A0A0L7QL68 E2C3B7 A0A0C9RDJ7 A0A151WFA5 A0A195BCZ4 F4WM91 A0A1W4X8F2 A0A195DCC6 A0A195F3M0 A0A158P2N3 A0A151IG74 A0A0M8ZZ82 A0A1B6D8D0 A0A1B6CP76 D6W756 A0A139WP70 A0A1B6CBE9 N6U6R4 A0A1B6EA89 A0A1Y1N7C8 A0A1J1J5N2 A0A1J1J1H5 A0A1J1J2K5 A0A0P8XYS8 B3MZ93 A0A0P8XIE8 A0A1B6EHJ6 A0A1B6MA26 A0A1B6LFK4 A0A1B6FFS5 A0A1B6F7U9 A0A1B6MU07 A0A1B6KZF9 A0A1B6KLT2 A0A0T6B8I8 A0A1W4WCS7 A0A1B6LBJ7 B3P9N6 A0A1B6I793 A0A1B6IVF4 A0A1B6IWC6 B4PW55 A0A1B6HKF1 A0A0R1EE50 A0A0R1E710 A0A0R1E7L8 B4IKS3 A0A0J9UT00 C3KGP2 A0A0J9S308 Q6WSQ9 H9XVQ2 A0A1L8DHX4 A0A1L8DHW9 A0A026W1Q3 B4NHH0 A0A221LED3 G3P7Y4 G3P7X1 G3P7Y0 A0A182VV94 A0A1I8PF41 A0A3B4XW15 A0A3B4UH05 A0A3B4UJQ2 A0A3B4DND5 A0A3Q2YU42 A0A3Q3NDX1 A0A0P7VLP6 A0A1W4XZ89
A0A088AUU6 A0A2A3E1E0 A0A154PKW9 A0A0C9R0A6 A0A0L7QL68 E2C3B7 A0A0C9RDJ7 A0A151WFA5 A0A195BCZ4 F4WM91 A0A1W4X8F2 A0A195DCC6 A0A195F3M0 A0A158P2N3 A0A151IG74 A0A0M8ZZ82 A0A1B6D8D0 A0A1B6CP76 D6W756 A0A139WP70 A0A1B6CBE9 N6U6R4 A0A1B6EA89 A0A1Y1N7C8 A0A1J1J5N2 A0A1J1J1H5 A0A1J1J2K5 A0A0P8XYS8 B3MZ93 A0A0P8XIE8 A0A1B6EHJ6 A0A1B6MA26 A0A1B6LFK4 A0A1B6FFS5 A0A1B6F7U9 A0A1B6MU07 A0A1B6KZF9 A0A1B6KLT2 A0A0T6B8I8 A0A1W4WCS7 A0A1B6LBJ7 B3P9N6 A0A1B6I793 A0A1B6IVF4 A0A1B6IWC6 B4PW55 A0A1B6HKF1 A0A0R1EE50 A0A0R1E710 A0A0R1E7L8 B4IKS3 A0A0J9UT00 C3KGP2 A0A0J9S308 Q6WSQ9 H9XVQ2 A0A1L8DHX4 A0A1L8DHW9 A0A026W1Q3 B4NHH0 A0A221LED3 G3P7Y4 G3P7X1 G3P7Y0 A0A182VV94 A0A1I8PF41 A0A3B4XW15 A0A3B4UH05 A0A3B4UJQ2 A0A3B4DND5 A0A3Q2YU42 A0A3Q3NDX1 A0A0P7VLP6 A0A1W4XZ89
EC Number
3.1.3.1
Pubmed
EMBL
NWSH01000008
PCG80959.1
KZ150133
PZC73031.1
RSAL01000181
RVE44921.1
+ More
KQ458671 KPJ05584.1 LADJ01030624 KPJ21204.1 AGBW02008142 OWR54109.1 KZ288455 PBC25518.1 KQ434954 KZC12521.1 GBYB01006367 JAG76134.1 KQ414934 KOC59357.1 GL452318 EFN77563.1 GBYB01006365 JAG76132.1 KQ983219 KYQ46518.1 KQ976522 KYM82060.1 GL888217 EGI64735.1 KQ980989 KYN10541.1 KQ981855 KYN34777.1 ADTU01001274 ADTU01001275 ADTU01001276 ADTU01001277 ADTU01001278 ADTU01001279 ADTU01001280 ADTU01001281 KQ977726 KYN00269.1 KQ435794 KOX74125.1 GEDC01015350 JAS21948.1 GEDC01022107 JAS15191.1 KQ971307 EFA11511.1 KYB29657.1 GEDC01026501 JAS10797.1 APGK01047235 KB741077 KB631792 ENN74257.1 ERL86116.1 GEDC01002450 JAS34848.1 GEZM01011184 JAV93609.1 CVRI01000066 CRL06193.1 CRL06192.1 CRL06194.1 CH902633 KPU74661.1 EDV33698.1 KPU74662.1 GECZ01032398 JAS37371.1 GEBQ01007192 JAT32785.1 GEBQ01017515 JAT22462.1 GECZ01020722 JAS49047.1 GECZ01023499 JAS46270.1 GEBQ01000592 JAT39385.1 GEBQ01023134 JAT16843.1 GEBQ01027584 JAT12393.1 LJIG01009142 KRT83672.1 GEBQ01019073 JAT20904.1 CH954184 EDV45199.1 GECU01024922 JAS82784.1 GECU01016787 JAS90919.1 GECU01016523 JAS91183.1 CM000161 EDW99360.1 GECU01032536 JAS75170.1 KRK05015.1 KRK05014.1 KRK05013.1 CH480855 EDW52663.1 CM002916 KMZ07416.1 BT082107 AE014135 ACQ45345.1 AFH06796.1 KMZ07415.1 AY271259 BT011052 AAQ01726.1 AAR31123.1 ABC65834.1 ABC65835.1 AFH06797.1 GFDF01008016 JAV06068.1 GFDF01008015 JAV06069.1 KK107488 EZA49958.1 CH964272 EDW83539.2 KY446553 ASM93515.1 JARO02001748 KPP74534.1
KQ458671 KPJ05584.1 LADJ01030624 KPJ21204.1 AGBW02008142 OWR54109.1 KZ288455 PBC25518.1 KQ434954 KZC12521.1 GBYB01006367 JAG76134.1 KQ414934 KOC59357.1 GL452318 EFN77563.1 GBYB01006365 JAG76132.1 KQ983219 KYQ46518.1 KQ976522 KYM82060.1 GL888217 EGI64735.1 KQ980989 KYN10541.1 KQ981855 KYN34777.1 ADTU01001274 ADTU01001275 ADTU01001276 ADTU01001277 ADTU01001278 ADTU01001279 ADTU01001280 ADTU01001281 KQ977726 KYN00269.1 KQ435794 KOX74125.1 GEDC01015350 JAS21948.1 GEDC01022107 JAS15191.1 KQ971307 EFA11511.1 KYB29657.1 GEDC01026501 JAS10797.1 APGK01047235 KB741077 KB631792 ENN74257.1 ERL86116.1 GEDC01002450 JAS34848.1 GEZM01011184 JAV93609.1 CVRI01000066 CRL06193.1 CRL06192.1 CRL06194.1 CH902633 KPU74661.1 EDV33698.1 KPU74662.1 GECZ01032398 JAS37371.1 GEBQ01007192 JAT32785.1 GEBQ01017515 JAT22462.1 GECZ01020722 JAS49047.1 GECZ01023499 JAS46270.1 GEBQ01000592 JAT39385.1 GEBQ01023134 JAT16843.1 GEBQ01027584 JAT12393.1 LJIG01009142 KRT83672.1 GEBQ01019073 JAT20904.1 CH954184 EDV45199.1 GECU01024922 JAS82784.1 GECU01016787 JAS90919.1 GECU01016523 JAS91183.1 CM000161 EDW99360.1 GECU01032536 JAS75170.1 KRK05015.1 KRK05014.1 KRK05013.1 CH480855 EDW52663.1 CM002916 KMZ07416.1 BT082107 AE014135 ACQ45345.1 AFH06796.1 KMZ07415.1 AY271259 BT011052 AAQ01726.1 AAR31123.1 ABC65834.1 ABC65835.1 AFH06797.1 GFDF01008016 JAV06068.1 GFDF01008015 JAV06069.1 KK107488 EZA49958.1 CH964272 EDW83539.2 KY446553 ASM93515.1 JARO02001748 KPP74534.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
UP000005203
+ More
UP000242457 UP000076502 UP000053825 UP000008237 UP000075809 UP000078540 UP000007755 UP000192223 UP000078492 UP000078541 UP000005205 UP000078542 UP000053105 UP000007266 UP000019118 UP000030742 UP000183832 UP000007801 UP000192221 UP000008711 UP000002282 UP000001292 UP000000803 UP000053097 UP000007798 UP000007635 UP000075920 UP000095300 UP000261360 UP000261420 UP000261440 UP000264820 UP000261640 UP000034805 UP000192224
UP000242457 UP000076502 UP000053825 UP000008237 UP000075809 UP000078540 UP000007755 UP000192223 UP000078492 UP000078541 UP000005205 UP000078542 UP000053105 UP000007266 UP000019118 UP000030742 UP000183832 UP000007801 UP000192221 UP000008711 UP000002282 UP000001292 UP000000803 UP000053097 UP000007798 UP000007635 UP000075920 UP000095300 UP000261360 UP000261420 UP000261440 UP000264820 UP000261640 UP000034805 UP000192224
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4KA52
A0A2W1BFJ0
A0A3S2TFY2
A0A194QJM4
A0A0N0PFG3
A0A212FK81
+ More
A0A088AUU6 A0A2A3E1E0 A0A154PKW9 A0A0C9R0A6 A0A0L7QL68 E2C3B7 A0A0C9RDJ7 A0A151WFA5 A0A195BCZ4 F4WM91 A0A1W4X8F2 A0A195DCC6 A0A195F3M0 A0A158P2N3 A0A151IG74 A0A0M8ZZ82 A0A1B6D8D0 A0A1B6CP76 D6W756 A0A139WP70 A0A1B6CBE9 N6U6R4 A0A1B6EA89 A0A1Y1N7C8 A0A1J1J5N2 A0A1J1J1H5 A0A1J1J2K5 A0A0P8XYS8 B3MZ93 A0A0P8XIE8 A0A1B6EHJ6 A0A1B6MA26 A0A1B6LFK4 A0A1B6FFS5 A0A1B6F7U9 A0A1B6MU07 A0A1B6KZF9 A0A1B6KLT2 A0A0T6B8I8 A0A1W4WCS7 A0A1B6LBJ7 B3P9N6 A0A1B6I793 A0A1B6IVF4 A0A1B6IWC6 B4PW55 A0A1B6HKF1 A0A0R1EE50 A0A0R1E710 A0A0R1E7L8 B4IKS3 A0A0J9UT00 C3KGP2 A0A0J9S308 Q6WSQ9 H9XVQ2 A0A1L8DHX4 A0A1L8DHW9 A0A026W1Q3 B4NHH0 A0A221LED3 G3P7Y4 G3P7X1 G3P7Y0 A0A182VV94 A0A1I8PF41 A0A3B4XW15 A0A3B4UH05 A0A3B4UJQ2 A0A3B4DND5 A0A3Q2YU42 A0A3Q3NDX1 A0A0P7VLP6 A0A1W4XZ89
A0A088AUU6 A0A2A3E1E0 A0A154PKW9 A0A0C9R0A6 A0A0L7QL68 E2C3B7 A0A0C9RDJ7 A0A151WFA5 A0A195BCZ4 F4WM91 A0A1W4X8F2 A0A195DCC6 A0A195F3M0 A0A158P2N3 A0A151IG74 A0A0M8ZZ82 A0A1B6D8D0 A0A1B6CP76 D6W756 A0A139WP70 A0A1B6CBE9 N6U6R4 A0A1B6EA89 A0A1Y1N7C8 A0A1J1J5N2 A0A1J1J1H5 A0A1J1J2K5 A0A0P8XYS8 B3MZ93 A0A0P8XIE8 A0A1B6EHJ6 A0A1B6MA26 A0A1B6LFK4 A0A1B6FFS5 A0A1B6F7U9 A0A1B6MU07 A0A1B6KZF9 A0A1B6KLT2 A0A0T6B8I8 A0A1W4WCS7 A0A1B6LBJ7 B3P9N6 A0A1B6I793 A0A1B6IVF4 A0A1B6IWC6 B4PW55 A0A1B6HKF1 A0A0R1EE50 A0A0R1E710 A0A0R1E7L8 B4IKS3 A0A0J9UT00 C3KGP2 A0A0J9S308 Q6WSQ9 H9XVQ2 A0A1L8DHX4 A0A1L8DHW9 A0A026W1Q3 B4NHH0 A0A221LED3 G3P7Y4 G3P7X1 G3P7Y0 A0A182VV94 A0A1I8PF41 A0A3B4XW15 A0A3B4UH05 A0A3B4UJQ2 A0A3B4DND5 A0A3Q2YU42 A0A3Q3NDX1 A0A0P7VLP6 A0A1W4XZ89
Ontologies
GO
PANTHER
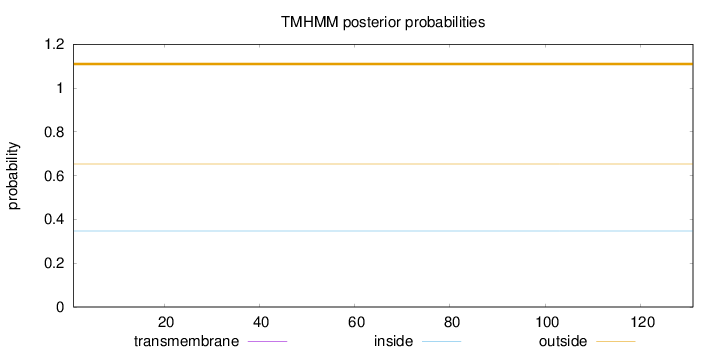
Topology
Length:
131
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.34718
outside
1 - 131
Population Genetic Test Statistics
Pi
169.594738
Theta
142.670381
Tajima's D
0.43534
CLR
0.6015
CSRT
0.493375331233438
Interpretation
Uncertain
