Pre Gene Modal
BGIBMGA004573
Annotation
PREDICTED:_probable_small_nuclear_ribonucleoprotein_G_[Papilio_polytes]
Full name
Small nuclear ribonucleoprotein G
+ More
Probable small nuclear ribonucleoprotein G
Probable small nuclear ribonucleoprotein G
Alternative Name
Sm protein G
Location in the cell
Cytoplasmic Reliability : 1.285 Mitochondrial Reliability : 1.34 Nuclear Reliability : 1.281
Sequence
CDS
ATGTCAAAAGCACATCCACCAGAATTGAAAAAATTCATGGACAAAAAATTGTCAATAAAGCTGAATGCGGGTCGCGCTGTTACCGGTGTGTTGCGCGGTTTTGATCCATTCATGAATTTAGTTCTTGATGAATCAGTCGAGGAGTGTAAAGACGGTCAGCGTAACAATGTTGGGATGGTGGTTAGTATATTTTCAAGAACACTCTAA
Protein
MSKAHPPELKKFMDKKLSIKLNAGRAVTGVLRGFDPFMNLVLDESVEECKDGQRNNVGMVVSIFSRTL
Summary
Description
Plays role in pre-mRNA splicing as core component of the SMN-Sm complex that mediates spliceosomal snRNP assembly (PubMed:18621711). Plays role in pre-mRNA splicing as component of the spliceosomal U1, U2, U4 and U5 small nuclear ribonucleoproteins (snRNPs), the building blocks of the spliceosome. Component of both the pre-catalytic spliceosome B complex and activated spliceosome C complexes. Is also a component of the minor U12 spliceosome (By similarity).
Plays role in pre-mRNA splicing as core component of the SMN-Sm complex that mediates spliceosomal snRNP assembly and as component of the spliceosomal U1, U2, U4 and U5 small nuclear ribonucleoproteins (snRNPs), the building blocks of the spliceosome. Component of both the pre-catalytic spliceosome B complex and activated spliceosome C complexes. Is also a component of the minor U12 spliceosome. As part of the U7 snRNP it is involved in histone 3'-end processing.
Plays role in pre-mRNA splicing as core component of the SMN-Sm complex that mediates spliceosomal snRNP assembly and as component of the spliceosomal U1, U2, U4 and U5 small nuclear ribonucleoproteins (snRNPs), the building blocks of the spliceosome. Component of both the pre-catalytic spliceosome B complex and activated spliceosome C complexes. Is also a component of the minor U12 spliceosome. As part of the U7 snRNP it is involved in histone 3'-end processing.
Subunit
Interacts with the SMN complex (PubMed:18621711). Core component of the spliceosomal U1, U2, U4 and U5 small nuclear ribonucleoproteins (snRNPs), the building blocks of the spliceosome. Most spliceosomal snRNPs contain a common set of Sm proteins, SNRPB, SNRPD1, SNRPD2, SNRPD3, SNRPE, SNRPF and SNRPG that assemble in a heptameric protein ring on the Sm site of the small nuclear RNA to form the core snRNP. Component of the U1 snRNP. Component of the U4/U6-U5 tri-snRNP complex. Component of the U7 snRNP complex. Component of the U11/U12 snRNPs that are part of the U12-type spliceosome (By similarity).
Similarity
Belongs to the snRNP Sm proteins family.
Keywords
Complete proteome
Cytoplasm
mRNA processing
mRNA splicing
Nucleus
Reference proteome
Ribonucleoprotein
RNA-binding
Spliceosome
Feature
chain Probable small nuclear ribonucleoprotein G
Uniprot
H9J4Y3
A0A212EMZ2
S4PGP0
A0A212F3N4
A0A2W1BMX0
A0A2A4KBB8
+ More
A0A2H1X0T2 Q1HQ15 A0A194QQ03 A0A0L7KIE3 E2B420 A0A2M4C589 A0A2M4AYN0 W5JQP2 A0A182FPH7 A0A182IRN9 A0A182XAE8 A0A1Q3F0J4 A0A182Y412 A0A182M220 A0A182QSQ0 A0A182UQA8 A0A182NR59 B0W6T2 A0A182VZV2 A0A182TEM8 Q7QCM2 A0A182RTI7 A0A182HS87 A0A232F0W0 K7J8R5 A0A1W4X4T0 J3JZA2 Q56FI6 A0A182S7C0 A0A182PL54 A0A182K3E1 Q16XZ1 A0A023EDW5 A0A2A3ECH3 A0A088AF01 E9ICB7 E2AS58 A0A0L7L418 A0A154PSX1 D6X2J3 A0A0L7QKV5 A0A0M8ZRW4 A0A026W947 F4X4B6 A0A158P3I9 A0A1B0CN65 T1H614 A0A1L8DGW6 A0A1W4V6C3 B4PXV5 B3MRC4 B3NVJ9 A0A1B0FJA6 A0A1I8M8R6 A0A1B0ALV8 A0A1I8PB85 A0A0L0CFR4 A0A1L8EI25 E0VUZ1 A0A3B0JCX6 B5DW06 B4G365 A0A2U9BZI3 B4R5V5 A0A1B2AJF9 B4IEQ4 Q9VXE0 A0A067R1E0 E9H7R1 A0A162QM09 U5EXK0 H3BEQ0 G3MKA9 R7UTD8 A0A2P8YLF4 B4MAP8 B4L7Y5 A0A1S3J1B5 A0A0P6GQN6 A0A0M5JB08 B4JK85 A0A224Z4M1 L7M3X4 A0A131XC56 B5M793 B4NEV7 A0A023GGT7 A0A1E1XTX9 A0A023FFY1 A0A023FW01 A0A1E1WYI2 A0A1D2NAZ6 A0A1J1IAH7 A0A2U4BYH4
A0A2H1X0T2 Q1HQ15 A0A194QQ03 A0A0L7KIE3 E2B420 A0A2M4C589 A0A2M4AYN0 W5JQP2 A0A182FPH7 A0A182IRN9 A0A182XAE8 A0A1Q3F0J4 A0A182Y412 A0A182M220 A0A182QSQ0 A0A182UQA8 A0A182NR59 B0W6T2 A0A182VZV2 A0A182TEM8 Q7QCM2 A0A182RTI7 A0A182HS87 A0A232F0W0 K7J8R5 A0A1W4X4T0 J3JZA2 Q56FI6 A0A182S7C0 A0A182PL54 A0A182K3E1 Q16XZ1 A0A023EDW5 A0A2A3ECH3 A0A088AF01 E9ICB7 E2AS58 A0A0L7L418 A0A154PSX1 D6X2J3 A0A0L7QKV5 A0A0M8ZRW4 A0A026W947 F4X4B6 A0A158P3I9 A0A1B0CN65 T1H614 A0A1L8DGW6 A0A1W4V6C3 B4PXV5 B3MRC4 B3NVJ9 A0A1B0FJA6 A0A1I8M8R6 A0A1B0ALV8 A0A1I8PB85 A0A0L0CFR4 A0A1L8EI25 E0VUZ1 A0A3B0JCX6 B5DW06 B4G365 A0A2U9BZI3 B4R5V5 A0A1B2AJF9 B4IEQ4 Q9VXE0 A0A067R1E0 E9H7R1 A0A162QM09 U5EXK0 H3BEQ0 G3MKA9 R7UTD8 A0A2P8YLF4 B4MAP8 B4L7Y5 A0A1S3J1B5 A0A0P6GQN6 A0A0M5JB08 B4JK85 A0A224Z4M1 L7M3X4 A0A131XC56 B5M793 B4NEV7 A0A023GGT7 A0A1E1XTX9 A0A023FFY1 A0A023FW01 A0A1E1WYI2 A0A1D2NAZ6 A0A1J1IAH7 A0A2U4BYH4
Pubmed
19121390
22118469
23622113
28756777
26354079
26227816
+ More
20798317 20920257 23761445 25244985 12364791 14747013 17210077 28648823 20075255 22516182 23537049 17510324 24945155 26483478 21282665 18362917 19820115 24508170 30249741 21719571 21347285 17994087 17550304 25315136 26108605 20566863 15632085 10731132 12537572 12537569 10908584 18621711 24845553 21292972 9215903 22216098 23254933 29403074 28797301 25576852 28049606 19320755 29209593 28503490 27289101
20798317 20920257 23761445 25244985 12364791 14747013 17210077 28648823 20075255 22516182 23537049 17510324 24945155 26483478 21282665 18362917 19820115 24508170 30249741 21719571 21347285 17994087 17550304 25315136 26108605 20566863 15632085 10731132 12537572 12537569 10908584 18621711 24845553 21292972 9215903 22216098 23254933 29403074 28797301 25576852 28049606 19320755 29209593 28503490 27289101
EMBL
BABH01025664
AGBW02013765
OWR42848.1
GAIX01006125
JAA86435.1
AGBW02010527
+ More
OWR48341.1 KZ150133 PZC73033.1 NWSH01000008 PCG80962.1 ODYU01012560 SOQ58955.1 DQ443237 ABF51326.1 KQ458671 KPJ05586.1 JTDY01009742 KOB62850.1 GL445463 EFN89534.1 GGFJ01011329 MBW60470.1 GGFK01012582 MBW45903.1 ADMH02000776 ETN65074.1 GFDL01013981 JAV21064.1 AXCM01000657 AXCN02001472 DS231849 EDS36913.1 AAAB01008859 EAA07689.2 APCN01000254 NNAY01001404 OXU24087.1 APGK01050286 BT128583 KB741169 KB632294 AEE63540.1 ENN73310.1 ERL91667.1 AY961514 AAX62416.1 CH477529 EAT39487.1 JXUM01098402 GAPW01006794 GAPW01006793 KQ564417 JAC06804.1 KXJ72321.1 KZ288287 PBC29437.1 GL762231 EFZ21774.1 GL442221 EFN63758.1 JTDY01003063 KOB70228.1 KQ435180 KZC15016.1 KQ971372 EFA09436.1 KQ414940 KOC59224.1 KQ435879 KOX69865.1 KK107323 QOIP01000002 EZA52580.1 RLU25776.1 GL888633 EGI58712.1 ADTU01008099 AJWK01019865 GFDF01008459 JAV05625.1 CM000162 EDX01941.1 CH902622 EDV34329.2 CH954180 EDV46391.1 CCAG010019526 JXJN01000167 JRES01000455 KNC31047.1 GFDG01000495 JAV18304.1 DS235797 EEB17197.1 OUUW01000005 SPP80234.1 CM000070 EDY68092.1 CH479179 EDW24260.1 CP026253 AWP09100.1 CM000366 EDX18105.1 KX531270 ANY27080.1 CH480832 EDW46158.1 AE014298 AY070698 AY113538 AAL48169.1 KK852781 KDR16631.1 GL732601 EFX72260.1 LRGB01000311 KZS19790.1 GANO01000914 JAB58957.1 AFYH01012359 AFYH01012360 JO842310 AEO33927.1 AMQN01006382 KB298218 ELU09428.1 PYGN01000512 PSN45044.1 CH940655 EDW66307.1 CH933814 EDW05560.1 GDIQ01042541 JAN52196.1 CP012528 ALC49974.1 CH916370 EDV99987.1 GFPF01011793 MAA22939.1 GACK01007235 JAA57799.1 GEFH01003347 JAP65234.1 EZ000280 ACG76261.1 CH964239 EDW82276.1 GBBM01003225 JAC32193.1 GFAA01000659 JAU02776.1 GBBK01004487 JAC19995.1 GBBL01002425 JAC24895.1 GFAC01007144 JAT92044.1 LJIJ01000112 ODN02423.1 CVRI01000047 CRK97219.1
OWR48341.1 KZ150133 PZC73033.1 NWSH01000008 PCG80962.1 ODYU01012560 SOQ58955.1 DQ443237 ABF51326.1 KQ458671 KPJ05586.1 JTDY01009742 KOB62850.1 GL445463 EFN89534.1 GGFJ01011329 MBW60470.1 GGFK01012582 MBW45903.1 ADMH02000776 ETN65074.1 GFDL01013981 JAV21064.1 AXCM01000657 AXCN02001472 DS231849 EDS36913.1 AAAB01008859 EAA07689.2 APCN01000254 NNAY01001404 OXU24087.1 APGK01050286 BT128583 KB741169 KB632294 AEE63540.1 ENN73310.1 ERL91667.1 AY961514 AAX62416.1 CH477529 EAT39487.1 JXUM01098402 GAPW01006794 GAPW01006793 KQ564417 JAC06804.1 KXJ72321.1 KZ288287 PBC29437.1 GL762231 EFZ21774.1 GL442221 EFN63758.1 JTDY01003063 KOB70228.1 KQ435180 KZC15016.1 KQ971372 EFA09436.1 KQ414940 KOC59224.1 KQ435879 KOX69865.1 KK107323 QOIP01000002 EZA52580.1 RLU25776.1 GL888633 EGI58712.1 ADTU01008099 AJWK01019865 GFDF01008459 JAV05625.1 CM000162 EDX01941.1 CH902622 EDV34329.2 CH954180 EDV46391.1 CCAG010019526 JXJN01000167 JRES01000455 KNC31047.1 GFDG01000495 JAV18304.1 DS235797 EEB17197.1 OUUW01000005 SPP80234.1 CM000070 EDY68092.1 CH479179 EDW24260.1 CP026253 AWP09100.1 CM000366 EDX18105.1 KX531270 ANY27080.1 CH480832 EDW46158.1 AE014298 AY070698 AY113538 AAL48169.1 KK852781 KDR16631.1 GL732601 EFX72260.1 LRGB01000311 KZS19790.1 GANO01000914 JAB58957.1 AFYH01012359 AFYH01012360 JO842310 AEO33927.1 AMQN01006382 KB298218 ELU09428.1 PYGN01000512 PSN45044.1 CH940655 EDW66307.1 CH933814 EDW05560.1 GDIQ01042541 JAN52196.1 CP012528 ALC49974.1 CH916370 EDV99987.1 GFPF01011793 MAA22939.1 GACK01007235 JAA57799.1 GEFH01003347 JAP65234.1 EZ000280 ACG76261.1 CH964239 EDW82276.1 GBBM01003225 JAC32193.1 GFAA01000659 JAU02776.1 GBBK01004487 JAC19995.1 GBBL01002425 JAC24895.1 GFAC01007144 JAT92044.1 LJIJ01000112 ODN02423.1 CVRI01000047 CRK97219.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000037510
UP000008237
+ More
UP000000673 UP000069272 UP000075880 UP000076407 UP000076408 UP000075883 UP000075886 UP000075903 UP000075884 UP000002320 UP000075920 UP000075902 UP000007062 UP000075900 UP000075840 UP000215335 UP000002358 UP000192223 UP000019118 UP000030742 UP000075901 UP000075885 UP000075881 UP000008820 UP000069940 UP000249989 UP000242457 UP000005203 UP000000311 UP000076502 UP000007266 UP000053825 UP000053105 UP000053097 UP000279307 UP000007755 UP000005205 UP000092461 UP000015102 UP000192221 UP000002282 UP000007801 UP000008711 UP000092444 UP000095301 UP000092460 UP000095300 UP000037069 UP000009046 UP000268350 UP000001819 UP000008744 UP000246464 UP000000304 UP000001292 UP000000803 UP000027135 UP000000305 UP000076858 UP000008672 UP000014760 UP000245037 UP000008792 UP000009192 UP000085678 UP000092553 UP000001070 UP000007798 UP000094527 UP000183832 UP000245320
UP000000673 UP000069272 UP000075880 UP000076407 UP000076408 UP000075883 UP000075886 UP000075903 UP000075884 UP000002320 UP000075920 UP000075902 UP000007062 UP000075900 UP000075840 UP000215335 UP000002358 UP000192223 UP000019118 UP000030742 UP000075901 UP000075885 UP000075881 UP000008820 UP000069940 UP000249989 UP000242457 UP000005203 UP000000311 UP000076502 UP000007266 UP000053825 UP000053105 UP000053097 UP000279307 UP000007755 UP000005205 UP000092461 UP000015102 UP000192221 UP000002282 UP000007801 UP000008711 UP000092444 UP000095301 UP000092460 UP000095300 UP000037069 UP000009046 UP000268350 UP000001819 UP000008744 UP000246464 UP000000304 UP000001292 UP000000803 UP000027135 UP000000305 UP000076858 UP000008672 UP000014760 UP000245037 UP000008792 UP000009192 UP000085678 UP000092553 UP000001070 UP000007798 UP000094527 UP000183832 UP000245320
Pfam
PF01423 LSM
SUPFAM
SSF50182
SSF50182
CDD
ProteinModelPortal
H9J4Y3
A0A212EMZ2
S4PGP0
A0A212F3N4
A0A2W1BMX0
A0A2A4KBB8
+ More
A0A2H1X0T2 Q1HQ15 A0A194QQ03 A0A0L7KIE3 E2B420 A0A2M4C589 A0A2M4AYN0 W5JQP2 A0A182FPH7 A0A182IRN9 A0A182XAE8 A0A1Q3F0J4 A0A182Y412 A0A182M220 A0A182QSQ0 A0A182UQA8 A0A182NR59 B0W6T2 A0A182VZV2 A0A182TEM8 Q7QCM2 A0A182RTI7 A0A182HS87 A0A232F0W0 K7J8R5 A0A1W4X4T0 J3JZA2 Q56FI6 A0A182S7C0 A0A182PL54 A0A182K3E1 Q16XZ1 A0A023EDW5 A0A2A3ECH3 A0A088AF01 E9ICB7 E2AS58 A0A0L7L418 A0A154PSX1 D6X2J3 A0A0L7QKV5 A0A0M8ZRW4 A0A026W947 F4X4B6 A0A158P3I9 A0A1B0CN65 T1H614 A0A1L8DGW6 A0A1W4V6C3 B4PXV5 B3MRC4 B3NVJ9 A0A1B0FJA6 A0A1I8M8R6 A0A1B0ALV8 A0A1I8PB85 A0A0L0CFR4 A0A1L8EI25 E0VUZ1 A0A3B0JCX6 B5DW06 B4G365 A0A2U9BZI3 B4R5V5 A0A1B2AJF9 B4IEQ4 Q9VXE0 A0A067R1E0 E9H7R1 A0A162QM09 U5EXK0 H3BEQ0 G3MKA9 R7UTD8 A0A2P8YLF4 B4MAP8 B4L7Y5 A0A1S3J1B5 A0A0P6GQN6 A0A0M5JB08 B4JK85 A0A224Z4M1 L7M3X4 A0A131XC56 B5M793 B4NEV7 A0A023GGT7 A0A1E1XTX9 A0A023FFY1 A0A023FW01 A0A1E1WYI2 A0A1D2NAZ6 A0A1J1IAH7 A0A2U4BYH4
A0A2H1X0T2 Q1HQ15 A0A194QQ03 A0A0L7KIE3 E2B420 A0A2M4C589 A0A2M4AYN0 W5JQP2 A0A182FPH7 A0A182IRN9 A0A182XAE8 A0A1Q3F0J4 A0A182Y412 A0A182M220 A0A182QSQ0 A0A182UQA8 A0A182NR59 B0W6T2 A0A182VZV2 A0A182TEM8 Q7QCM2 A0A182RTI7 A0A182HS87 A0A232F0W0 K7J8R5 A0A1W4X4T0 J3JZA2 Q56FI6 A0A182S7C0 A0A182PL54 A0A182K3E1 Q16XZ1 A0A023EDW5 A0A2A3ECH3 A0A088AF01 E9ICB7 E2AS58 A0A0L7L418 A0A154PSX1 D6X2J3 A0A0L7QKV5 A0A0M8ZRW4 A0A026W947 F4X4B6 A0A158P3I9 A0A1B0CN65 T1H614 A0A1L8DGW6 A0A1W4V6C3 B4PXV5 B3MRC4 B3NVJ9 A0A1B0FJA6 A0A1I8M8R6 A0A1B0ALV8 A0A1I8PB85 A0A0L0CFR4 A0A1L8EI25 E0VUZ1 A0A3B0JCX6 B5DW06 B4G365 A0A2U9BZI3 B4R5V5 A0A1B2AJF9 B4IEQ4 Q9VXE0 A0A067R1E0 E9H7R1 A0A162QM09 U5EXK0 H3BEQ0 G3MKA9 R7UTD8 A0A2P8YLF4 B4MAP8 B4L7Y5 A0A1S3J1B5 A0A0P6GQN6 A0A0M5JB08 B4JK85 A0A224Z4M1 L7M3X4 A0A131XC56 B5M793 B4NEV7 A0A023GGT7 A0A1E1XTX9 A0A023FFY1 A0A023FW01 A0A1E1WYI2 A0A1D2NAZ6 A0A1J1IAH7 A0A2U4BYH4
PDB
6QX9
E-value=1.09086e-21,
Score=248
Ontologies
GO
Topology
Subcellular location
Nucleus
Cytoplasm SMN-mediated assembly into core snRNPs occurs in the cytosol before SMN-mediated transport to the nucleus to be included in spliceosomes. With evidence from 1 publications.
Cytosol SMN-mediated assembly into core snRNPs occurs in the cytosol before SMN-mediated transport to the nucleus to be included in spliceosomes. With evidence from 1 publications.
Cytoplasm SMN-mediated assembly into core snRNPs occurs in the cytosol before SMN-mediated transport to the nucleus to be included in spliceosomes. With evidence from 1 publications.
Cytosol SMN-mediated assembly into core snRNPs occurs in the cytosol before SMN-mediated transport to the nucleus to be included in spliceosomes. With evidence from 1 publications.
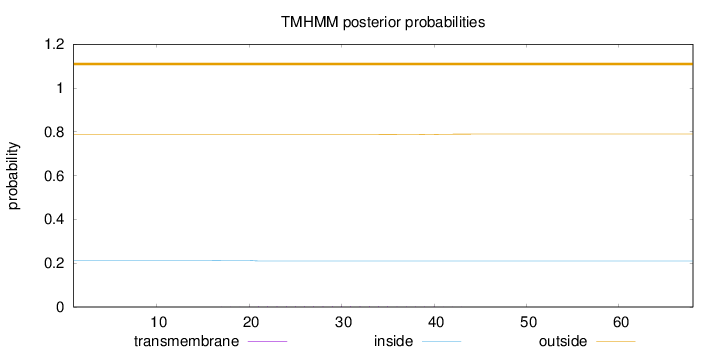
Length:
68
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05328
Exp number, first 60 AAs:
0.05328
Total prob of N-in:
0.21275
outside
1 - 68
Population Genetic Test Statistics
Pi
66.520574
Theta
105.984053
Tajima's D
-1.167971
CLR
279.451386
CSRT
0.107394630268487
Interpretation
Uncertain
