Gene
KWMTBOMO16077
Pre Gene Modal
BGIBMGA004574
Annotation
PREDICTED:_solute_carrier_family_12_member_9_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.642
Sequence
CDS
ATGATTGATACTTTTTTTGTACCCCAGCTGAAAGAAGTGGATGTTGACGAGTCAGGTTATTTAGTAGGTAATGCCGGTCTTCTGGTGACATTAGCGCAATTCGCCGTGGCGTACCTCATAGTGCTGTTCACGGTGACCTCAATATGTGCGATATCGACAAACGGGGCGGTCGAAGGAGGCGGCGTGTATTTCATGTTGAGCAGAACCCTAGGTCCCGAATTCGGAGGTGCAATAGGGACGTTGTTCTTCTTCGCGAATGTGGTGTCCAGTGCTTTGTGCATATCAGCGTGCACTGAGGCCTTGGTTGAAAACTTCGGAACCAATGGGTACTTAGTGGGTGAGAGTGGTGGTATCCCGGACAGCTGGTGGTACCGCTTCCTGTATCGCTCCGTGCTGAATGCGGTCGGGCTGGGTGTGTCCCTCGCGGGCGCCTCGCTGTTCGCCAGGACCAGCCTCGCCATCTGGGTCACGATGCTGGTCTGCTTAGCCAGCGCCTTCTTGAGCTTCTTCATCACGAGTCCTGCGCAGATCGAGAAGCCAGATTCGAATCACTTAATAAACACGACGTATCTGAACTACACCGGGCTGAGCGTGTCGACGCTGGCTTCGAACATGTACCCGCAGTACGGGCGCGACTACACAACGGCGAGCGGGCAGCTCGTGGACTTCGCGTCGGTGCTGGGCGTGCTGTTCACCGGCGTCACCGGCGTCATGGCCGGCGCCAACATGAGCGGGGAGTTGAAGCACCCGTCGCGCAGTATCCCGCGCGGCACGCTCGCGGCGCTGCTGTTCACGGCCGCGTCCTACGCCGCGCTGAGCCTGCTGACCGCGGCCACGTGCTCCAGGGAGCTGCTGCAGAACAACTACGTGTACCTGCTGCCCATCAACGTGTGGCCACCCTTCATAGCCGTCGGTATGATAGTAGCTACATTTTCGGCCGCACTCTCCAACCTGATCGGCGCTTCTAGAGTGTTGGAAGCGTTGGCCAAAGATGACATATTTGGTTTCCTTCTTCGTCCGATGGTGTCGCGTTCCGGGAACCCTGTGCTGGCGGTGGTGGTGTCCTGGGCGCTGGTGCAGGTGGGCGTGGCGGCGGACTCCCTCAACGCCATAGCGCAAGTCAACTCGGTCCTCTTCATGACGAGCTATTTCGCCATCAATCTGGCTTGCTTGGGACTGGAGCTCGCTTCGGCTCCTAACTTCAGGCCGACGTTCCGTCAGTTCTCGTGGGCGACGTCGCTGGCGGGGCTGTGCGGCAGCGCGGCGCTGATGTTCTCGCTGCGCGCGCTGTACGCGGCGGGCGGCGCGCTGGCGGCCGCGGGGCTGGTGCTGGCGCTGCACCTGCTGTCGCCCGCCGCCGCCGAGCCCAAGTGGGGCAGCCTCAGCCAGGCGCTCATCTTCCACCAGGTCCGCAAGTACCTGCTGCTTCTGGACCCGCGGCGGGAGCACGTCAAGTTCTGGCGGCCGCAGATGCTGCTGCTGGTAGCCTCGCCGAGACAGTCCGCGCCGCTCATAGACTTCGTCAACGACCAGAAGAAGGGCGGGCTGTTCGTGCTGGGCCACGTGCGCGTGGGGCGGCTGGACGGCGCAGGGGACCCGCTCGCGGAGGAGCACCAGTACTGGCTGAAGCTCATCGACCATCTCAAGGTGAAGGCGTTCGTGGAGCTGTGCCTGGCGGAGTCGGTGCGCAGCGGCGCCGCCCACCTGGCGCGCCTGTCCGGTCTGGGCGCCATGAAGCCCGACACCGTGATGCTCGGCTTCTGGGACGACGCGCAGCCGCGGGACTTCTTCAGGGAACCGGAGTCGCCGTACCGCACGGAGCTGTTCGACGCGGGCGGCGGCGCCTTCCCCGCCCGCCGCGCCGGCGCCGCGCGCCTGCCGCCCGCCGAGTACGTGCGCATACTGTCCGACGTGCTCTGCGTCAACAAGAACGTGTGCATATGCAGGCACTTCCATCTGCTGGATATGGCGGCGGTCGAACGTAAGTCCCCGAGGTTGAAGTACATCGACGTGTGGCTGGTGGATCTGCTGGCGCCGAGCCGCGAGGAGGCGTTCTCGGTGCGGGGGCTGTTCGCGCTGCAGCTGGCGGCCGTGGTGCGGTCCGCGCGCGGCTGGCAACACCTGCAACTCCGCGTGCACACCACCAGACACGCAAGCCTGACCGTGGGCTCCGGCGACAGTCCGGCGCCGGACGGGCCGCCGCTGCAGCAGCGCCTCGAGGAGCTCCTAAAGCTGCTGCGGATCAATGCCACTACGCACATCATCTCAGATTGGCCGAAGCTGGACCTGAGCAGGTGGTCCGATACGGACGAGGACCTGTACCAGCGGGTCCCCGCCACCTACCTCAAGACCGTGAACAGTATAGTCCGGGAGCGCTGCGAGCAGGGCACCGCCGTGACCTTCGTCCAGCTTAGCGCCCCGCCCCGCGAGGCCGACGAGCTGTGCGAGCAGTACCTGCAGGTGCTGGACGAATTCACGAAGGATTTATCGCCGACGATTCTAGTGAGGGGCTTAAAGTCGGTCACGTCCACGTCTCTGTAA
Protein
MIDTFFVPQLKEVDVDESGYLVGNAGLLVTLAQFAVAYLIVLFTVTSICAISTNGAVEGGGVYFMLSRTLGPEFGGAIGTLFFFANVVSSALCISACTEALVENFGTNGYLVGESGGIPDSWWYRFLYRSVLNAVGLGVSLAGASLFARTSLAIWVTMLVCLASAFLSFFITSPAQIEKPDSNHLINTTYLNYTGLSVSTLASNMYPQYGRDYTTASGQLVDFASVLGVLFTGVTGVMAGANMSGELKHPSRSIPRGTLAALLFTAASYAALSLLTAATCSRELLQNNYVYLLPINVWPPFIAVGMIVATFSAALSNLIGASRVLEALAKDDIFGFLLRPMVSRSGNPVLAVVVSWALVQVGVAADSLNAIAQVNSVLFMTSYFAINLACLGLELASAPNFRPTFRQFSWATSLAGLCGSAALMFSLRALYAAGGALAAAGLVLALHLLSPAAAEPKWGSLSQALIFHQVRKYLLLLDPRREHVKFWRPQMLLLVASPRQSAPLIDFVNDQKKGGLFVLGHVRVGRLDGAGDPLAEEHQYWLKLIDHLKVKAFVELCLAESVRSGAAHLARLSGLGAMKPDTVMLGFWDDAQPRDFFREPESPYRTELFDAGGGAFPARRAGAARLPPAEYVRILSDVLCVNKNVCICRHFHLLDMAAVERKSPRLKYIDVWLVDLLAPSREEAFSVRGLFALQLAAVVRSARGWQHLQLRVHTTRHASLTVGSGDSPAPDGPPLQQRLEELLKLLRINATTHIISDWPKLDLSRWSDTDEDLYQRVPATYLKTVNSIVRERCEQGTAVTFVQLSAPPREADELCEQYLQVLDEFTKDLSPTILVRGLKSVTSTSL
Summary
Cofactor
Mn(2+)
Similarity
Belongs to the glycosyltransferase 2 family. GalNAc-T subfamily.
Uniprot
A0A2W1B7E2
A0A1E1W9H6
A0A026X0M0
E2BRE5
E2AV35
A0A195FJ83
+ More
F4WUB9 A0A195C7H7 A0A151J6F0 A0A310SFS2 A0A087ZXW7 A0A154P0U6 A0A0J7KYF9 A0A151I4T2 A0A2A3EB55 A0A023EW80 A0A182H5M6 Q16K72 A0A1S4FYI0 W8AP20 A0A0K8UF73 A0A084W5J7 A0A2J7RFJ2 A0A182QR84 A0A182MUA1 A0A182YHG0 A0A182K2J1 A0A1A9XDN1 A0A067RLG4 A0A1Q3FDF0 A0A1Q3FCX5 A0A1Q3FDH9 A0A1I8NU70 A0A182N8T8 A0A1Q3FDK6 B0XBU1 Q7Q0N1 A0A182JCB5 A0A2M3ZF02 A0A0L0CHT8 A0A2M4BCI2 A0A2M4BCL8 W5JVY1 A0A1L8DF00 A0A2M4A833 A0A2M4A887 A0A182FMY1 U5ENZ6 A0A182VSV1 A0A0A9W9C4 A0A1B0DF86 A0A146M6A1 A0A1W4XW25 A0A0M8ZUT7 A0A1A9W0D8 D6WJT4 A0A182RAC2 A0A182X7B0 A0A182KJV3 A0A182I0T6 A0A1I8N2L1 A0A0C9RBD0 A0A182VNW4 A0A1W4V9S5 A0A0V0G4G4 A0A069DZM1 A0A182PK75 A0A224XJG4 A0A2S2PQA5 A0A0J9R419 Q9VJ75 A0A0K8W3V6 A0A0K8U9Y9 A0A2S2Q248 B4P9V3 B3NLC5 A0A2H8TGP2 A0A1J1HZ59 B4I5H9 Q16K71 E0VL18 Q29MP1 B4KK24 J9JR18 A0A0M4E5U5 A0A3B0K6A6 B4LTJ4 A0A1Y1K8K4 B3MM29 A0A336LS50 B4JDM5 B4MZF7 A0A0K8SW94 A0A2M3ZKU9 A0A1B0CEI6 A0A1D2NK86 A0A1Q3FDH2 A0A1Q3FDC1
F4WUB9 A0A195C7H7 A0A151J6F0 A0A310SFS2 A0A087ZXW7 A0A154P0U6 A0A0J7KYF9 A0A151I4T2 A0A2A3EB55 A0A023EW80 A0A182H5M6 Q16K72 A0A1S4FYI0 W8AP20 A0A0K8UF73 A0A084W5J7 A0A2J7RFJ2 A0A182QR84 A0A182MUA1 A0A182YHG0 A0A182K2J1 A0A1A9XDN1 A0A067RLG4 A0A1Q3FDF0 A0A1Q3FCX5 A0A1Q3FDH9 A0A1I8NU70 A0A182N8T8 A0A1Q3FDK6 B0XBU1 Q7Q0N1 A0A182JCB5 A0A2M3ZF02 A0A0L0CHT8 A0A2M4BCI2 A0A2M4BCL8 W5JVY1 A0A1L8DF00 A0A2M4A833 A0A2M4A887 A0A182FMY1 U5ENZ6 A0A182VSV1 A0A0A9W9C4 A0A1B0DF86 A0A146M6A1 A0A1W4XW25 A0A0M8ZUT7 A0A1A9W0D8 D6WJT4 A0A182RAC2 A0A182X7B0 A0A182KJV3 A0A182I0T6 A0A1I8N2L1 A0A0C9RBD0 A0A182VNW4 A0A1W4V9S5 A0A0V0G4G4 A0A069DZM1 A0A182PK75 A0A224XJG4 A0A2S2PQA5 A0A0J9R419 Q9VJ75 A0A0K8W3V6 A0A0K8U9Y9 A0A2S2Q248 B4P9V3 B3NLC5 A0A2H8TGP2 A0A1J1HZ59 B4I5H9 Q16K71 E0VL18 Q29MP1 B4KK24 J9JR18 A0A0M4E5U5 A0A3B0K6A6 B4LTJ4 A0A1Y1K8K4 B3MM29 A0A336LS50 B4JDM5 B4MZF7 A0A0K8SW94 A0A2M3ZKU9 A0A1B0CEI6 A0A1D2NK86 A0A1Q3FDH2 A0A1Q3FDC1
Pubmed
28756777
24508170
30249741
20798317
21719571
24945155
+ More
26483478 17519023 17510324 24495485 24438588 25244985 24845553 12364791 14747013 17210077 26108605 20920257 23761445 25401762 26823975 18362917 19820115 20966253 25315136 26334808 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 20566863 15632085 28004739 27289101
26483478 17519023 17510324 24495485 24438588 25244985 24845553 12364791 14747013 17210077 26108605 20920257 23761445 25401762 26823975 18362917 19820115 20966253 25315136 26334808 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 20566863 15632085 28004739 27289101
EMBL
KZ150249
PZC71839.1
GDQN01007431
JAT83623.1
KK107054
QOIP01000004
+ More
EZA61551.1 RLU23582.1 GL449962 EFN81723.1 GL443013 EFN62702.1 KQ981522 KYN40720.1 GL888355 EGI62227.1 KQ978143 KYM96797.1 KQ979854 KYN18797.1 KQ761729 OAD57161.1 KQ434796 KZC05559.1 LBMM01002020 KMQ95381.1 KQ976445 KYM86103.1 KZ288308 PBC28704.1 GAPW01000138 JAC13460.1 JXUM01112102 KQ565562 KXJ70879.1 EF173370 CH477974 ABM68597.1 EAT34694.1 GAMC01018793 GAMC01018791 JAB87762.1 GDHF01027098 JAI25216.1 ATLV01020668 KE525304 KFB45491.1 NEVH01004413 PNF39599.1 AXCN02001866 AXCM01000224 KK852439 KDR23883.1 GFDL01009475 JAV25570.1 GFDL01009611 JAV25434.1 GFDL01009438 JAV25607.1 GFDL01009399 JAV25646.1 DS232653 EDS44470.1 AAAB01008980 EAA13952.3 GGFM01006338 MBW27089.1 JRES01000359 KNC31953.1 GGFJ01001614 MBW50755.1 GGFJ01001629 MBW50770.1 ADMH02000247 ETN67250.1 GFDF01009154 JAV04930.1 GGFK01003618 MBW36939.1 GGFK01003629 MBW36950.1 GANO01000381 JAB59490.1 GBHO01042149 JAG01455.1 AJVK01058606 GDHC01003346 JAQ15283.1 KQ435834 KOX71575.1 KQ971343 EFA04772.2 APCN01005264 GBYB01012684 GBYB01012688 GBYB01013779 JAG82451.1 JAG82455.1 JAG83546.1 GECL01003192 JAP02932.1 GBGD01000400 JAC88489.1 GFTR01008255 JAW08171.1 GGMR01019033 MBY31652.1 CM002910 KMY90838.1 AE014134 AY047570 AAF53679.1 AAK77302.1 GDHF01006592 JAI45722.1 GDHF01028916 JAI23398.1 GGMS01002654 MBY71857.1 CM000158 EDW90294.1 CH954179 EDV54775.1 GFXV01001460 MBW13265.1 CVRI01000037 CRK93243.1 CH480822 EDW55635.1 EAT34695.1 DS235263 EEB14074.1 CH379060 EAL33652.1 CH933807 EDW11542.1 ABLF02027289 CP012523 ALC39558.1 OUUW01000004 SPP78998.1 CH940649 EDW64967.1 GEZM01089085 JAV57724.1 CH902620 EDV30844.1 UFQT01000144 SSX20842.1 CH916368 EDW03395.1 CH963920 EDW77742.1 GBRD01008656 JAG57165.1 GGFM01008374 MBW29125.1 AJWK01008903 AJWK01008904 AJWK01008905 LJIJ01000021 ODN05525.1 GFDL01009431 JAV25614.1 GFDL01009526 JAV25519.1
EZA61551.1 RLU23582.1 GL449962 EFN81723.1 GL443013 EFN62702.1 KQ981522 KYN40720.1 GL888355 EGI62227.1 KQ978143 KYM96797.1 KQ979854 KYN18797.1 KQ761729 OAD57161.1 KQ434796 KZC05559.1 LBMM01002020 KMQ95381.1 KQ976445 KYM86103.1 KZ288308 PBC28704.1 GAPW01000138 JAC13460.1 JXUM01112102 KQ565562 KXJ70879.1 EF173370 CH477974 ABM68597.1 EAT34694.1 GAMC01018793 GAMC01018791 JAB87762.1 GDHF01027098 JAI25216.1 ATLV01020668 KE525304 KFB45491.1 NEVH01004413 PNF39599.1 AXCN02001866 AXCM01000224 KK852439 KDR23883.1 GFDL01009475 JAV25570.1 GFDL01009611 JAV25434.1 GFDL01009438 JAV25607.1 GFDL01009399 JAV25646.1 DS232653 EDS44470.1 AAAB01008980 EAA13952.3 GGFM01006338 MBW27089.1 JRES01000359 KNC31953.1 GGFJ01001614 MBW50755.1 GGFJ01001629 MBW50770.1 ADMH02000247 ETN67250.1 GFDF01009154 JAV04930.1 GGFK01003618 MBW36939.1 GGFK01003629 MBW36950.1 GANO01000381 JAB59490.1 GBHO01042149 JAG01455.1 AJVK01058606 GDHC01003346 JAQ15283.1 KQ435834 KOX71575.1 KQ971343 EFA04772.2 APCN01005264 GBYB01012684 GBYB01012688 GBYB01013779 JAG82451.1 JAG82455.1 JAG83546.1 GECL01003192 JAP02932.1 GBGD01000400 JAC88489.1 GFTR01008255 JAW08171.1 GGMR01019033 MBY31652.1 CM002910 KMY90838.1 AE014134 AY047570 AAF53679.1 AAK77302.1 GDHF01006592 JAI45722.1 GDHF01028916 JAI23398.1 GGMS01002654 MBY71857.1 CM000158 EDW90294.1 CH954179 EDV54775.1 GFXV01001460 MBW13265.1 CVRI01000037 CRK93243.1 CH480822 EDW55635.1 EAT34695.1 DS235263 EEB14074.1 CH379060 EAL33652.1 CH933807 EDW11542.1 ABLF02027289 CP012523 ALC39558.1 OUUW01000004 SPP78998.1 CH940649 EDW64967.1 GEZM01089085 JAV57724.1 CH902620 EDV30844.1 UFQT01000144 SSX20842.1 CH916368 EDW03395.1 CH963920 EDW77742.1 GBRD01008656 JAG57165.1 GGFM01008374 MBW29125.1 AJWK01008903 AJWK01008904 AJWK01008905 LJIJ01000021 ODN05525.1 GFDL01009431 JAV25614.1 GFDL01009526 JAV25519.1
Proteomes
UP000053097
UP000279307
UP000008237
UP000000311
UP000078541
UP000007755
+ More
UP000078542 UP000078492 UP000005203 UP000076502 UP000036403 UP000078540 UP000242457 UP000069940 UP000249989 UP000008820 UP000030765 UP000235965 UP000075886 UP000075883 UP000076408 UP000075881 UP000092443 UP000027135 UP000095300 UP000075884 UP000002320 UP000007062 UP000075880 UP000037069 UP000000673 UP000069272 UP000075920 UP000092462 UP000192223 UP000053105 UP000091820 UP000007266 UP000075900 UP000076407 UP000075882 UP000075840 UP000095301 UP000075903 UP000192221 UP000075885 UP000000803 UP000002282 UP000008711 UP000183832 UP000001292 UP000009046 UP000001819 UP000009192 UP000007819 UP000092553 UP000268350 UP000008792 UP000007801 UP000001070 UP000007798 UP000092461 UP000094527
UP000078542 UP000078492 UP000005203 UP000076502 UP000036403 UP000078540 UP000242457 UP000069940 UP000249989 UP000008820 UP000030765 UP000235965 UP000075886 UP000075883 UP000076408 UP000075881 UP000092443 UP000027135 UP000095300 UP000075884 UP000002320 UP000007062 UP000075880 UP000037069 UP000000673 UP000069272 UP000075920 UP000092462 UP000192223 UP000053105 UP000091820 UP000007266 UP000075900 UP000076407 UP000075882 UP000075840 UP000095301 UP000075903 UP000192221 UP000075885 UP000000803 UP000002282 UP000008711 UP000183832 UP000001292 UP000009046 UP000001819 UP000009192 UP000007819 UP000092553 UP000268350 UP000008792 UP000007801 UP000001070 UP000007798 UP000092461 UP000094527
PRIDE
Pfam
Interpro
IPR018491
SLC12_C
+ More
IPR004841 AA-permease/SLC12A_dom
IPR007858 Dpy-30_motif
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR018170 Aldo/ket_reductase_CS
IPR001680 WD40_repeat
IPR006692 Coatomer_WD-assoc_reg
IPR023210 NADP_OxRdtase_dom
IPR010714 Coatomer_asu_C
IPR036322 WD40_repeat_dom_sf
IPR036812 NADP_OxRdtase_dom_sf
IPR017986 WD40_repeat_dom
IPR020471 Aldo/keto_reductase
IPR001173 Glyco_trans_2-like
IPR000772 Ricin_B_lectin
IPR035992 Ricin_B-like_lectins
IPR029044 Nucleotide-diphossugar_trans
IPR004841 AA-permease/SLC12A_dom
IPR007858 Dpy-30_motif
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR018170 Aldo/ket_reductase_CS
IPR001680 WD40_repeat
IPR006692 Coatomer_WD-assoc_reg
IPR023210 NADP_OxRdtase_dom
IPR010714 Coatomer_asu_C
IPR036322 WD40_repeat_dom_sf
IPR036812 NADP_OxRdtase_dom_sf
IPR017986 WD40_repeat_dom
IPR020471 Aldo/keto_reductase
IPR001173 Glyco_trans_2-like
IPR000772 Ricin_B_lectin
IPR035992 Ricin_B-like_lectins
IPR029044 Nucleotide-diphossugar_trans
Gene 3D
ProteinModelPortal
A0A2W1B7E2
A0A1E1W9H6
A0A026X0M0
E2BRE5
E2AV35
A0A195FJ83
+ More
F4WUB9 A0A195C7H7 A0A151J6F0 A0A310SFS2 A0A087ZXW7 A0A154P0U6 A0A0J7KYF9 A0A151I4T2 A0A2A3EB55 A0A023EW80 A0A182H5M6 Q16K72 A0A1S4FYI0 W8AP20 A0A0K8UF73 A0A084W5J7 A0A2J7RFJ2 A0A182QR84 A0A182MUA1 A0A182YHG0 A0A182K2J1 A0A1A9XDN1 A0A067RLG4 A0A1Q3FDF0 A0A1Q3FCX5 A0A1Q3FDH9 A0A1I8NU70 A0A182N8T8 A0A1Q3FDK6 B0XBU1 Q7Q0N1 A0A182JCB5 A0A2M3ZF02 A0A0L0CHT8 A0A2M4BCI2 A0A2M4BCL8 W5JVY1 A0A1L8DF00 A0A2M4A833 A0A2M4A887 A0A182FMY1 U5ENZ6 A0A182VSV1 A0A0A9W9C4 A0A1B0DF86 A0A146M6A1 A0A1W4XW25 A0A0M8ZUT7 A0A1A9W0D8 D6WJT4 A0A182RAC2 A0A182X7B0 A0A182KJV3 A0A182I0T6 A0A1I8N2L1 A0A0C9RBD0 A0A182VNW4 A0A1W4V9S5 A0A0V0G4G4 A0A069DZM1 A0A182PK75 A0A224XJG4 A0A2S2PQA5 A0A0J9R419 Q9VJ75 A0A0K8W3V6 A0A0K8U9Y9 A0A2S2Q248 B4P9V3 B3NLC5 A0A2H8TGP2 A0A1J1HZ59 B4I5H9 Q16K71 E0VL18 Q29MP1 B4KK24 J9JR18 A0A0M4E5U5 A0A3B0K6A6 B4LTJ4 A0A1Y1K8K4 B3MM29 A0A336LS50 B4JDM5 B4MZF7 A0A0K8SW94 A0A2M3ZKU9 A0A1B0CEI6 A0A1D2NK86 A0A1Q3FDH2 A0A1Q3FDC1
F4WUB9 A0A195C7H7 A0A151J6F0 A0A310SFS2 A0A087ZXW7 A0A154P0U6 A0A0J7KYF9 A0A151I4T2 A0A2A3EB55 A0A023EW80 A0A182H5M6 Q16K72 A0A1S4FYI0 W8AP20 A0A0K8UF73 A0A084W5J7 A0A2J7RFJ2 A0A182QR84 A0A182MUA1 A0A182YHG0 A0A182K2J1 A0A1A9XDN1 A0A067RLG4 A0A1Q3FDF0 A0A1Q3FCX5 A0A1Q3FDH9 A0A1I8NU70 A0A182N8T8 A0A1Q3FDK6 B0XBU1 Q7Q0N1 A0A182JCB5 A0A2M3ZF02 A0A0L0CHT8 A0A2M4BCI2 A0A2M4BCL8 W5JVY1 A0A1L8DF00 A0A2M4A833 A0A2M4A887 A0A182FMY1 U5ENZ6 A0A182VSV1 A0A0A9W9C4 A0A1B0DF86 A0A146M6A1 A0A1W4XW25 A0A0M8ZUT7 A0A1A9W0D8 D6WJT4 A0A182RAC2 A0A182X7B0 A0A182KJV3 A0A182I0T6 A0A1I8N2L1 A0A0C9RBD0 A0A182VNW4 A0A1W4V9S5 A0A0V0G4G4 A0A069DZM1 A0A182PK75 A0A224XJG4 A0A2S2PQA5 A0A0J9R419 Q9VJ75 A0A0K8W3V6 A0A0K8U9Y9 A0A2S2Q248 B4P9V3 B3NLC5 A0A2H8TGP2 A0A1J1HZ59 B4I5H9 Q16K71 E0VL18 Q29MP1 B4KK24 J9JR18 A0A0M4E5U5 A0A3B0K6A6 B4LTJ4 A0A1Y1K8K4 B3MM29 A0A336LS50 B4JDM5 B4MZF7 A0A0K8SW94 A0A2M3ZKU9 A0A1B0CEI6 A0A1D2NK86 A0A1Q3FDH2 A0A1Q3FDC1
Ontologies
GO
GO:0016021
GO:0055085
GO:0006811
GO:0005215
GO:1902476
GO:0005623
GO:0015379
GO:0055064
GO:0006884
GO:0055075
GO:1990573
GO:0005198
GO:0016192
GO:0016491
GO:0006886
GO:0030126
GO:0030246
GO:0016757
GO:0006486
GO:0005794
GO:0016020
GO:0006810
GO:0022857
GO:0001678
GO:0005536
GO:0046835
GO:0006281
GO:0006259
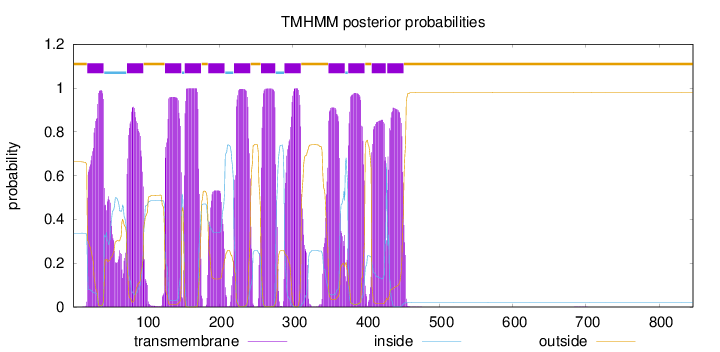
Topology
Length:
846
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
245.5721
Exp number, first 60 AAs:
25.75424
Total prob of N-in:
0.33718
POSSIBLE N-term signal
sequence
outside
1 - 19
TMhelix
20 - 42
inside
43 - 73
TMhelix
74 - 96
outside
97 - 125
TMhelix
126 - 148
inside
149 - 152
TMhelix
153 - 175
outside
176 - 184
TMhelix
185 - 207
inside
208 - 219
TMhelix
220 - 242
outside
243 - 256
TMhelix
257 - 276
inside
277 - 288
TMhelix
289 - 311
outside
312 - 348
TMhelix
349 - 371
inside
372 - 375
TMhelix
376 - 398
outside
399 - 407
TMhelix
408 - 427
inside
428 - 428
TMhelix
429 - 451
outside
452 - 846
Population Genetic Test Statistics
Pi
326.027348
Theta
159.31821
Tajima's D
3.497042
CLR
0.257665
CSRT
0.994550272486376
Interpretation
Uncertain
