Gene
KWMTBOMO16073
Pre Gene Modal
BGIBMGA004576
Annotation
PREDICTED:_uncharacterized_protein_LOC106713509_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.44 Mitochondrial Reliability : 1.424 Peroxisomal Reliability : 1.154
Sequence
CDS
ATGTGGCGTTTTGGTAAGAAAAACCTTTACGTCTGGAGTCTTGAAACCACGACGCTGATAAAAGTACTGGACGCTCACTTTGGCAGAATAGTCCAACTGGAACCGTTGACTGTTGGAGCTTGGAACAATGTGATAACGTCGTCCATAGACAGGACGGTCAAGATATGGAACATTGATAACATATTTGAGCAAGTACATAAGATTGATCGACAAGAACTGCCCATTGATTCTATTAGCCTAGCACGCGACAAGCAACTAGCGGTAACCGTCACCCGTAGCTGTTGGGGCCTCTGGGATACGAGCACCGGTCGGTTGATGGCGCAATTAGCTGACGCTCCGCTCGGCGCCATCGTCACGCACGCGGTCATCACGCCCACTGGAGATCACGTAGTCACTGCGGAATCGGGCTGCGTTATTATATGGGATACTCCTGAAAAACAGGGAATCTTCAAGGAAGAACAAAAAGGTGTAAAACAAGTTCTGCTTCTAGAAGACGGGACGAAATTCATCACGATATCTATGCAGGTGCCCGCTGATAACACGGATAACGTTGCTGTTGCTTATATCGTGGCAAGAACTATACCTGACGGTGAAAGAATTTACACAGCAGAATACGACGTTCGGGGTACCGGCTACAGAGCCGCTGTGGTTTCTGCAGAGGAACATCATCTAGTTGCTCCCGGAGTGGACAAGTCCTCCAGGGACTGTCTACATGTATTCCATGCTAGAACCGGAGCGAGGATACATAGGATACCAATTAAGAACAGCGGCATCAAAGACATGCAATCTATTGTGGCGTTGCCCCACAAGGCGCTCTGGGTGGCAGTCGTCGGTAATGACAAAGCCGGTATATTGGACATCAAAAGCAAAAGACTCATCAGATTTGTTCCACGTTGGAATGGCGCATGTACTCGCGATGGTAAGCTGGGGCTCTGTGCGCCTTCCAGAGGGGGGCTCGACCTCATAGAACTAAAGAGAGGAACATCGGTGCACCCCCTTATATACAGGGCGGCTGAAGGCGTATTTTCCATCATCACCATGTTTAGCTCGACAGACCATTATGTATTTTATTATCACAGCGGCAGGAAAACACTGAGAGTTTTCAGAACAATAGACGGTCGTGCTATAGCTGAGTACCGGGCGCCGGCCGAAGTCACAGGCGTGGCCAGCGCTCATGGAGGCAAAGCTGTCGCTATCGCCTCACAGGACGGTTGTCTCACAATCCTTAACATTGTTGACCCATACGAATGA
Protein
MWRFGKKNLYVWSLETTTLIKVLDAHFGRIVQLEPLTVGAWNNVITSSIDRTVKIWNIDNIFEQVHKIDRQELPIDSISLARDKQLAVTVTRSCWGLWDTSTGRLMAQLADAPLGAIVTHAVITPTGDHVVTAESGCVIIWDTPEKQGIFKEEQKGVKQVLLLEDGTKFITISMQVPADNTDNVAVAYIVARTIPDGERIYTAEYDVRGTGYRAAVVSAEEHHLVAPGVDKSSRDCLHVFHARTGARIHRIPIKNSGIKDMQSIVALPHKALWVAVVGNDKAGILDIKSKRLIRFVPRWNGACTRDGKLGLCAPSRGGLDLIELKRGTSVHPLIYRAAEGVFSIITMFSSTDHYVFYYHSGRKTLRVFRTIDGRAIAEYRAPAEVTGVASAHGGKAVAIASQDGCLTILNIVDPYE
Summary
Uniprot
A0A2A4JCC0
A0A2W1BJX8
A0A2H1W6D7
A0A194R440
A0A3S2TKP3
A0A212F853
+ More
A0A0N1IE15 A0A2J7RFG5 A0A2H8TJ14 A0A195FJX0 A0A158NRE9 F4WAY8 A0A195BFH5 A0A151JBR8 A0A0N0U7I8 A0A3L8DKH5 E2AUX2 A0A151IFS8 E0VJC5 A0A151XF31 A0A2A3ED85 A0A026WF60 A0A088AR94 A0A182SYU0 A0A182WDI4 A0A310SMH0 A0A1W4XMN4 A0A2S2PDS7 J9JUM0 E2BLG4 A0A182LYL0 A0A1B0CWF8 A0A182RTD4 A0A182N2C0 A0A182U632 A0A182HWC7 A0A182VE12 T1I544 A0A034V0T7 Q7QHF1 A0A182YB27 A0A182NZL9 A0A336MEJ1 A0A182L3H2 A0A182QN31 A0A182JNC1 A0A182X5M2 W5J7I9 A0A182J713 A0A084VHT9 A0A232F7U5 A0A182FKK4 Q0IFD7 B0WL71 A0A0L7RCH5 A0A067QH37 A0A182G340 N6TQT6 U4UIZ9 A0A1Q3FSW4 A0A0K8VJN8 A0A0L0BP11 A0A1I8N5Z0 A0A1W4VEG6 B4LY14 B3MXB4 A0A0K8WB69 B3P0X9 Q9VEZ2 B4PR45 B4QXQ8 B4JGA1 B4NL15 B4KAF4 A0A1I8NYU7 A0A1I8NYT7 A0A1D2N487 A0A3B0JTE2 Q294U8 A0A0R3NQ94 B4GML1 A0A0R3NNX5 A0A0R3NJD4 A0A1B0GDT5 A0A1A9X452 A0A1A9V657 A0A1A9X8M2 A0A1A9ZY44 A0A1B0BF52 A0A0M4EGP6 A0A226DAE0 A0A2P8XE62 K7JQM9 E9HFF2 A0A0P5WGQ2 A0A0P6HBY7 A0A0P5TR82 A0A162R3C6 A0A0P5UKF9
A0A0N1IE15 A0A2J7RFG5 A0A2H8TJ14 A0A195FJX0 A0A158NRE9 F4WAY8 A0A195BFH5 A0A151JBR8 A0A0N0U7I8 A0A3L8DKH5 E2AUX2 A0A151IFS8 E0VJC5 A0A151XF31 A0A2A3ED85 A0A026WF60 A0A088AR94 A0A182SYU0 A0A182WDI4 A0A310SMH0 A0A1W4XMN4 A0A2S2PDS7 J9JUM0 E2BLG4 A0A182LYL0 A0A1B0CWF8 A0A182RTD4 A0A182N2C0 A0A182U632 A0A182HWC7 A0A182VE12 T1I544 A0A034V0T7 Q7QHF1 A0A182YB27 A0A182NZL9 A0A336MEJ1 A0A182L3H2 A0A182QN31 A0A182JNC1 A0A182X5M2 W5J7I9 A0A182J713 A0A084VHT9 A0A232F7U5 A0A182FKK4 Q0IFD7 B0WL71 A0A0L7RCH5 A0A067QH37 A0A182G340 N6TQT6 U4UIZ9 A0A1Q3FSW4 A0A0K8VJN8 A0A0L0BP11 A0A1I8N5Z0 A0A1W4VEG6 B4LY14 B3MXB4 A0A0K8WB69 B3P0X9 Q9VEZ2 B4PR45 B4QXQ8 B4JGA1 B4NL15 B4KAF4 A0A1I8NYU7 A0A1I8NYT7 A0A1D2N487 A0A3B0JTE2 Q294U8 A0A0R3NQ94 B4GML1 A0A0R3NNX5 A0A0R3NJD4 A0A1B0GDT5 A0A1A9X452 A0A1A9V657 A0A1A9X8M2 A0A1A9ZY44 A0A1B0BF52 A0A0M4EGP6 A0A226DAE0 A0A2P8XE62 K7JQM9 E9HFF2 A0A0P5WGQ2 A0A0P6HBY7 A0A0P5TR82 A0A162R3C6 A0A0P5UKF9
Pubmed
28756777
26354079
22118469
21347285
21719571
30249741
+ More
20798317 20566863 24508170 25348373 12364791 14747013 17210077 25244985 20966253 20920257 23761445 24438588 28648823 17510324 24845553 26483478 23537049 26108605 25315136 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 27289101 15632085 29403074 20075255 21292972
20798317 20566863 24508170 25348373 12364791 14747013 17210077 25244985 20966253 20920257 23761445 24438588 28648823 17510324 24845553 26483478 23537049 26108605 25315136 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 27289101 15632085 29403074 20075255 21292972
EMBL
NWSH01001908
PCG69725.1
KZ149997
PZC75382.1
ODYU01006321
SOQ48064.1
+ More
KQ460761 KPJ12573.1 RSAL01000078 RVE48741.1 AGBW02009801 OWR49898.1 KQ458842 KPJ04911.1 NEVH01004413 PNF39578.1 GFXV01001877 MBW13682.1 KQ981522 KYN40557.1 ADTU01023849 GL888056 EGI68608.1 KQ976504 KYM82920.1 KQ979122 KYN22530.1 KQ435711 KOX79674.1 QOIP01000007 RLU20944.1 GL442898 EFN62780.1 KQ977771 KYM99979.1 DS235222 EEB13481.1 KQ982205 KYQ58996.1 KZ288289 PBC29252.1 KK107238 EZA54682.1 KQ763581 OAD54968.1 GGMR01014988 MBY27607.1 ABLF02037162 GL449017 EFN83516.1 AXCM01001359 AJWK01032340 AJWK01032341 AJWK01032342 AJWK01032343 AJWK01032344 AJWK01032345 APCN01001484 ACPB03010524 GAKP01023219 JAC35737.1 AAAB01008816 EAA05136.3 UFQT01001083 SSX28802.1 AXCN02001729 ADMH02002090 ETN59363.1 ATLV01013235 KE524847 KFB37533.1 NNAY01000701 OXU26911.1 CH477356 EAT42778.1 DS231980 EDS30244.1 KQ414615 KOC68524.1 KK853768 KDQ84119.1 JXUM01140736 JXUM01140737 JXUM01140738 JXUM01140739 KQ569115 KXJ68757.1 APGK01055161 APGK01055162 KB741261 ENN71565.1 KB632344 ERL93057.1 GFDL01004390 JAV30655.1 GDHF01013227 JAI39087.1 JRES01001582 KNC21746.1 CH940650 EDW66880.1 CH902629 EDV35337.1 GDHF01004214 JAI48100.1 CH954181 EDV49098.1 AE014297 AAF55270.1 CM000160 EDW97367.1 CM000364 EDX12732.1 CH916369 EDV92570.1 CH964272 EDW84218.1 CH933806 EDW15667.1 LJIJ01000234 ODN00079.1 OUUW01000008 SPP83672.1 CM000070 EAL28866.2 KRT01136.1 CH479185 EDW38085.1 KRT01135.1 KRT01137.1 CCAG010012025 JXJN01013283 JXJN01013284 JXJN01013285 CP012526 ALC45346.1 LNIX01000028 OXA41697.1 PYGN01002564 PSN30262.1 GL732635 EFX69546.1 GDIP01086821 JAM16894.1 GDIQ01022359 JAN72378.1 GDIP01123917 JAL79797.1 LRGB01000243 KZS20194.1 GDIP01111618 JAL92096.1
KQ460761 KPJ12573.1 RSAL01000078 RVE48741.1 AGBW02009801 OWR49898.1 KQ458842 KPJ04911.1 NEVH01004413 PNF39578.1 GFXV01001877 MBW13682.1 KQ981522 KYN40557.1 ADTU01023849 GL888056 EGI68608.1 KQ976504 KYM82920.1 KQ979122 KYN22530.1 KQ435711 KOX79674.1 QOIP01000007 RLU20944.1 GL442898 EFN62780.1 KQ977771 KYM99979.1 DS235222 EEB13481.1 KQ982205 KYQ58996.1 KZ288289 PBC29252.1 KK107238 EZA54682.1 KQ763581 OAD54968.1 GGMR01014988 MBY27607.1 ABLF02037162 GL449017 EFN83516.1 AXCM01001359 AJWK01032340 AJWK01032341 AJWK01032342 AJWK01032343 AJWK01032344 AJWK01032345 APCN01001484 ACPB03010524 GAKP01023219 JAC35737.1 AAAB01008816 EAA05136.3 UFQT01001083 SSX28802.1 AXCN02001729 ADMH02002090 ETN59363.1 ATLV01013235 KE524847 KFB37533.1 NNAY01000701 OXU26911.1 CH477356 EAT42778.1 DS231980 EDS30244.1 KQ414615 KOC68524.1 KK853768 KDQ84119.1 JXUM01140736 JXUM01140737 JXUM01140738 JXUM01140739 KQ569115 KXJ68757.1 APGK01055161 APGK01055162 KB741261 ENN71565.1 KB632344 ERL93057.1 GFDL01004390 JAV30655.1 GDHF01013227 JAI39087.1 JRES01001582 KNC21746.1 CH940650 EDW66880.1 CH902629 EDV35337.1 GDHF01004214 JAI48100.1 CH954181 EDV49098.1 AE014297 AAF55270.1 CM000160 EDW97367.1 CM000364 EDX12732.1 CH916369 EDV92570.1 CH964272 EDW84218.1 CH933806 EDW15667.1 LJIJ01000234 ODN00079.1 OUUW01000008 SPP83672.1 CM000070 EAL28866.2 KRT01136.1 CH479185 EDW38085.1 KRT01135.1 KRT01137.1 CCAG010012025 JXJN01013283 JXJN01013284 JXJN01013285 CP012526 ALC45346.1 LNIX01000028 OXA41697.1 PYGN01002564 PSN30262.1 GL732635 EFX69546.1 GDIP01086821 JAM16894.1 GDIQ01022359 JAN72378.1 GDIP01123917 JAL79797.1 LRGB01000243 KZS20194.1 GDIP01111618 JAL92096.1
Proteomes
UP000218220
UP000053240
UP000283053
UP000007151
UP000053268
UP000235965
+ More
UP000078541 UP000005205 UP000007755 UP000078540 UP000078492 UP000053105 UP000279307 UP000000311 UP000078542 UP000009046 UP000075809 UP000242457 UP000053097 UP000005203 UP000075901 UP000075920 UP000192223 UP000007819 UP000008237 UP000075883 UP000092461 UP000075900 UP000075884 UP000075902 UP000075840 UP000075903 UP000015103 UP000007062 UP000076408 UP000075885 UP000075882 UP000075886 UP000075881 UP000076407 UP000000673 UP000075880 UP000030765 UP000215335 UP000069272 UP000008820 UP000002320 UP000053825 UP000027135 UP000069940 UP000249989 UP000019118 UP000030742 UP000037069 UP000095301 UP000192221 UP000008792 UP000007801 UP000008711 UP000000803 UP000002282 UP000000304 UP000001070 UP000007798 UP000009192 UP000095300 UP000094527 UP000268350 UP000001819 UP000008744 UP000092444 UP000091820 UP000078200 UP000092443 UP000092445 UP000092460 UP000092553 UP000198287 UP000245037 UP000002358 UP000000305 UP000076858
UP000078541 UP000005205 UP000007755 UP000078540 UP000078492 UP000053105 UP000279307 UP000000311 UP000078542 UP000009046 UP000075809 UP000242457 UP000053097 UP000005203 UP000075901 UP000075920 UP000192223 UP000007819 UP000008237 UP000075883 UP000092461 UP000075900 UP000075884 UP000075902 UP000075840 UP000075903 UP000015103 UP000007062 UP000076408 UP000075885 UP000075882 UP000075886 UP000075881 UP000076407 UP000000673 UP000075880 UP000030765 UP000215335 UP000069272 UP000008820 UP000002320 UP000053825 UP000027135 UP000069940 UP000249989 UP000019118 UP000030742 UP000037069 UP000095301 UP000192221 UP000008792 UP000007801 UP000008711 UP000000803 UP000002282 UP000000304 UP000001070 UP000007798 UP000009192 UP000095300 UP000094527 UP000268350 UP000001819 UP000008744 UP000092444 UP000091820 UP000078200 UP000092443 UP000092445 UP000092460 UP000092553 UP000198287 UP000245037 UP000002358 UP000000305 UP000076858
PRIDE
Interpro
IPR011047
Quinoprotein_ADH-like_supfam
+ More
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR011048 Haem_d1_sf
IPR019775 WD40_repeat_CS
IPR027417 P-loop_NTPase
IPR025139 DUF4062
IPR007111 NACHT_NTPase
IPR011044 Quino_amine_DH_bsu
IPR036322 WD40_repeat_dom_sf
IPR041664 AAA_16
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR011048 Haem_d1_sf
IPR019775 WD40_repeat_CS
IPR027417 P-loop_NTPase
IPR025139 DUF4062
IPR007111 NACHT_NTPase
IPR011044 Quino_amine_DH_bsu
IPR036322 WD40_repeat_dom_sf
IPR041664 AAA_16
SUPFAM
Gene 3D
ProteinModelPortal
A0A2A4JCC0
A0A2W1BJX8
A0A2H1W6D7
A0A194R440
A0A3S2TKP3
A0A212F853
+ More
A0A0N1IE15 A0A2J7RFG5 A0A2H8TJ14 A0A195FJX0 A0A158NRE9 F4WAY8 A0A195BFH5 A0A151JBR8 A0A0N0U7I8 A0A3L8DKH5 E2AUX2 A0A151IFS8 E0VJC5 A0A151XF31 A0A2A3ED85 A0A026WF60 A0A088AR94 A0A182SYU0 A0A182WDI4 A0A310SMH0 A0A1W4XMN4 A0A2S2PDS7 J9JUM0 E2BLG4 A0A182LYL0 A0A1B0CWF8 A0A182RTD4 A0A182N2C0 A0A182U632 A0A182HWC7 A0A182VE12 T1I544 A0A034V0T7 Q7QHF1 A0A182YB27 A0A182NZL9 A0A336MEJ1 A0A182L3H2 A0A182QN31 A0A182JNC1 A0A182X5M2 W5J7I9 A0A182J713 A0A084VHT9 A0A232F7U5 A0A182FKK4 Q0IFD7 B0WL71 A0A0L7RCH5 A0A067QH37 A0A182G340 N6TQT6 U4UIZ9 A0A1Q3FSW4 A0A0K8VJN8 A0A0L0BP11 A0A1I8N5Z0 A0A1W4VEG6 B4LY14 B3MXB4 A0A0K8WB69 B3P0X9 Q9VEZ2 B4PR45 B4QXQ8 B4JGA1 B4NL15 B4KAF4 A0A1I8NYU7 A0A1I8NYT7 A0A1D2N487 A0A3B0JTE2 Q294U8 A0A0R3NQ94 B4GML1 A0A0R3NNX5 A0A0R3NJD4 A0A1B0GDT5 A0A1A9X452 A0A1A9V657 A0A1A9X8M2 A0A1A9ZY44 A0A1B0BF52 A0A0M4EGP6 A0A226DAE0 A0A2P8XE62 K7JQM9 E9HFF2 A0A0P5WGQ2 A0A0P6HBY7 A0A0P5TR82 A0A162R3C6 A0A0P5UKF9
A0A0N1IE15 A0A2J7RFG5 A0A2H8TJ14 A0A195FJX0 A0A158NRE9 F4WAY8 A0A195BFH5 A0A151JBR8 A0A0N0U7I8 A0A3L8DKH5 E2AUX2 A0A151IFS8 E0VJC5 A0A151XF31 A0A2A3ED85 A0A026WF60 A0A088AR94 A0A182SYU0 A0A182WDI4 A0A310SMH0 A0A1W4XMN4 A0A2S2PDS7 J9JUM0 E2BLG4 A0A182LYL0 A0A1B0CWF8 A0A182RTD4 A0A182N2C0 A0A182U632 A0A182HWC7 A0A182VE12 T1I544 A0A034V0T7 Q7QHF1 A0A182YB27 A0A182NZL9 A0A336MEJ1 A0A182L3H2 A0A182QN31 A0A182JNC1 A0A182X5M2 W5J7I9 A0A182J713 A0A084VHT9 A0A232F7U5 A0A182FKK4 Q0IFD7 B0WL71 A0A0L7RCH5 A0A067QH37 A0A182G340 N6TQT6 U4UIZ9 A0A1Q3FSW4 A0A0K8VJN8 A0A0L0BP11 A0A1I8N5Z0 A0A1W4VEG6 B4LY14 B3MXB4 A0A0K8WB69 B3P0X9 Q9VEZ2 B4PR45 B4QXQ8 B4JGA1 B4NL15 B4KAF4 A0A1I8NYU7 A0A1I8NYT7 A0A1D2N487 A0A3B0JTE2 Q294U8 A0A0R3NQ94 B4GML1 A0A0R3NNX5 A0A0R3NJD4 A0A1B0GDT5 A0A1A9X452 A0A1A9V657 A0A1A9X8M2 A0A1A9ZY44 A0A1B0BF52 A0A0M4EGP6 A0A226DAE0 A0A2P8XE62 K7JQM9 E9HFF2 A0A0P5WGQ2 A0A0P6HBY7 A0A0P5TR82 A0A162R3C6 A0A0P5UKF9
PDB
6J6Q
E-value=0.000442876,
Score=103
Ontologies
GO
Topology
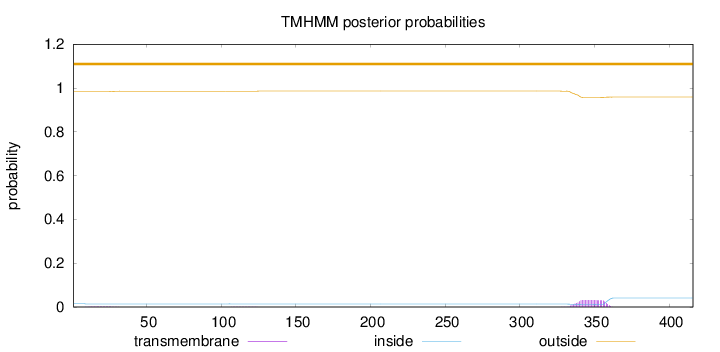
Length:
416
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.71391
Exp number, first 60 AAs:
0.0279
Total prob of N-in:
0.01547
outside
1 - 416
Population Genetic Test Statistics
Pi
306.830286
Theta
164.466183
Tajima's D
3.467589
CLR
0.364735
CSRT
0.993200339983001
Interpretation
Uncertain
