Pre Gene Modal
BGIBMGA004570
Annotation
AF141930_1_high_affinity_GABA_transporter_[Trichoplusia_ni]
Full name
Transporter
Location in the cell
PlasmaMembrane Reliability : 4.912
Sequence
CDS
ATGGACGTGAAAAACGATGATATTGAACTTAGCGTGCAGGGAACAAGCAAGCCAAGTGACGTCGCGATCAAATCCAATTTGCCGGAGCGAGAATCCTGGGCCAGCAAGCTAGACTTCATCCTTTCTGTAGTTGGCTTGGCCATTGGACTTGGTAACGTTTGGCGTTTCCCTTACCTCTGTTACAAGAATGGCGGGGGAGCCTTCCTCATTCCTTACTTCCTTACACTCTTCCTCGCTGGTATACCGATGTTTTTCATGGAACTGGCCATGGGACAAATGCTGACGATTGGAGGCCTCGGCGTTTTTAAGATCGCCCCGATTTTTAAAGGCATTGGCTACGCTGCAGCCGTGATGTCCTGTTGGATGAATGTTTACTACATCGTAATTCTGGCCTGGGCTATATTTTACTTCTTCATGTCGATGCGTTCAGATGTCCCTTGGCGTAATTGCGACAATTACTGGAACACACCGAGTTGCGTGAACCCTTACGATCGTAAGAATCTGACTTGCTGGTCTAGCAAGGATATGTCGACTTATTGCATCCTCAACGGCAAAAACTTCACCAAAGCTATGCTCTCTGATCCGGTCAAGGAATTTTGGGAACGCCGCGCCCTACAGATTTCATCAGGCATCGAGCATATTGGCAACATCCGCTGGGAATTAGCCGGGACCTTGCTTCTCGTTTGGGTTCTGTGTTACTTCTGCATTTGGAAAGGAGTGAGATGGACAGGCAAAGTTGTTTACTTTACTGCACTGTTTCCATACTTTTTGCTTACCGTGCTGTTAATTAGAGGTATAACGTTACCGGGAGCCTTGGAAGGGATAAAGTTCTACGTGATGCCAAACATGTCCAAGCTACTGGAGTCTGAAGTATGGATTGACGCTGTCACTCAAATCTTCTTCTCGTACGGATTAGGCCTGGGCACTTTAGTAGCTCTTGGTAGCTACAACAAGTTCACTAATAATGTTTACAAGGACGCTCTGATAGTCTGCTCTGTTAACTCCAGCACGTCAATGTTTGCTGGTTTCGTCATCTTTTCTGTGGTTGGCTTTATGGCGCATGAACAGCAGCGTCCTGTTGCTGAAGTGGCCGCTTCTGGTCCGGGCTTGGCATTTCTGGCGTACCCATCAGCCGTCCTGCAGTTGCCAGGTGCCCCGCTCTGGTCTTGCCTGTTCTTCTTCATGCTACTCCTCATTGGCTTGGACAGCCAGTTCTGCACCATGGAAGGCTTCGTGACCGCCGTCATCGACGAATGGCCTAAACTCTTGAGAAGGCGAAAGGAAGTGTTCATTGCAATTACTTGCGTTATTTCTTACCTGGTCGGATTGTCTTGTATCTCTGAAGGTGGCATGTACGTTTTTCAAATCCTCGACTCGTACGCTGTCTCTGGTTTCTGTCTGCTGTTCCTCATTTTCTTTGAGTGCGTCTCCATCTCGTGGGCCTTCGGTGTTAACCGTTTCTATGACGGGATCAAGGAAATGATCGGATACTACCCAATGATTTGGTGGAAGTTCTGCTGGGTTGGATTCACTCCTGCCATTTGCATTAGCGTTTTCATCTTCAATCTCGTGCAGTGGACTCCAATCAAATACATGAATTACGAATACCCCTGGTGGTCCCACGCATTCGGATGGTTTACCGCTTTATCTTCGATGTTGTGTATTCCTGGGTACATGGTGTACCTGTGGCGTGTCACCCCCGGAACCTGGCATGAGAAATTCAACACGATCGTACGCATTCCCGAAGACGTGCCATCTCTACGAACAAAGATGCAAGCTGAAGAACAAGCAAAACATGGCAAAGCTTAG
Protein
MDVKNDDIELSVQGTSKPSDVAIKSNLPERESWASKLDFILSVVGLAIGLGNVWRFPYLCYKNGGGAFLIPYFLTLFLAGIPMFFMELAMGQMLTIGGLGVFKIAPIFKGIGYAAAVMSCWMNVYYIVILAWAIFYFFMSMRSDVPWRNCDNYWNTPSCVNPYDRKNLTCWSSKDMSTYCILNGKNFTKAMLSDPVKEFWERRALQISSGIEHIGNIRWELAGTLLLVWVLCYFCIWKGVRWTGKVVYFTALFPYFLLTVLLIRGITLPGALEGIKFYVMPNMSKLLESEVWIDAVTQIFFSYGLGLGTLVALGSYNKFTNNVYKDALIVCSVNSSTSMFAGFVIFSVVGFMAHEQQRPVAEVAASGPGLAFLAYPSAVLQLPGAPLWSCLFFFMLLLIGLDSQFCTMEGFVTAVIDEWPKLLRRRKEVFIAITCVISYLVGLSCISEGGMYVFQILDSYAVSGFCLLFLIFFECVSISWAFGVNRFYDGIKEMIGYYPMIWWKFCWVGFTPAICISVFIFNLVQWTPIKYMNYEYPWWSHAFGWFTALSSMLCIPGYMVYLWRVTPGTWHEKFNTIVRIPEDVPSLRTKMQAEEQAKHGKA
Summary
Similarity
Belongs to the sodium:neurotransmitter symporter (SNF) (TC 2.A.22) family.
Uniprot
Q9NJI0
A0A0N0PA64
A0A2W1BJM4
A0A2A4JHY8
A0A0U4ZU62
A0A0G3VIB4
+ More
Q25512 A0A212F833 A0A336L7S6 A0A336MIE5 D6WKL1 A0A1Q3FPR1 E0VAA4 A0A1Y1LKL2 K7IX81 A0A1A9WQD1 E2BL27 A0A0K8U2S1 A0A034WRI5 A0A0C9RDY6 A0A1B0BZS7 A0A1A9YA80 A0A0A1WUC9 A0A026WPT1 B4H889 F4WH84 A0A0M9AD94 A0A158NPD8 A0A088AT93 A0A151X6C7 A0A151IH15 A0A2A3EIS8 B4NHM9 A0A1W4V4Y7 A0A195E530 A0A0K8UT91 A0A0L7R7Y6 A0A1A9ZJY6 E2A4G9 A0A3B0K464 A0A195F4N9 T1I4T4 W8C964 B4LF59 A0A0N7Z8U8 A0A023F218 A0A1W4VH63 B3MZY1 A0A0J7L5P3 A0A067R6S2 B4KVK2 B4J3B5 A0A0A9WTG2 A0A0L0CI82 B4IIX8 B3P9S2 A0A0J9S2H0 B4PW19 A0A1B6GXH5 A0A1B6JT61 Q9V4E7 A0A2J7R419 A0A1B6CA44 Q6NND1 A0A310SS41 A0A2P8YR78 A0A0N1IJP4 A0A1B6KZ53 A0A1B6LMH6 A0A1J1J9R4 Q20C86 A0A1A9VFK0 A0A2H1X1A9 A0A1B0G9X9 A0A182F2W0 W5J7P6 E2AQ81 B0X5H6 A0A182NBS6 A0A026WWC1 A0A182R5I3 A0A182J5D0 A0A182KU05 A0A182KGN3 Q16S73 A0A151XIU1 A0A154PBJ8 A0A182Y0Q0 A0A195FMW4 A0A182MDI6 A0A182HRW0 E9IDG2 A0A2A3EDC6 A0A1S4FRB9 A0A1Q3FQ46 Q6TLC7 A0A1Q3FQ74 A0A084WTZ6 A0A182GV19 Q6TLC6
Q25512 A0A212F833 A0A336L7S6 A0A336MIE5 D6WKL1 A0A1Q3FPR1 E0VAA4 A0A1Y1LKL2 K7IX81 A0A1A9WQD1 E2BL27 A0A0K8U2S1 A0A034WRI5 A0A0C9RDY6 A0A1B0BZS7 A0A1A9YA80 A0A0A1WUC9 A0A026WPT1 B4H889 F4WH84 A0A0M9AD94 A0A158NPD8 A0A088AT93 A0A151X6C7 A0A151IH15 A0A2A3EIS8 B4NHM9 A0A1W4V4Y7 A0A195E530 A0A0K8UT91 A0A0L7R7Y6 A0A1A9ZJY6 E2A4G9 A0A3B0K464 A0A195F4N9 T1I4T4 W8C964 B4LF59 A0A0N7Z8U8 A0A023F218 A0A1W4VH63 B3MZY1 A0A0J7L5P3 A0A067R6S2 B4KVK2 B4J3B5 A0A0A9WTG2 A0A0L0CI82 B4IIX8 B3P9S2 A0A0J9S2H0 B4PW19 A0A1B6GXH5 A0A1B6JT61 Q9V4E7 A0A2J7R419 A0A1B6CA44 Q6NND1 A0A310SS41 A0A2P8YR78 A0A0N1IJP4 A0A1B6KZ53 A0A1B6LMH6 A0A1J1J9R4 Q20C86 A0A1A9VFK0 A0A2H1X1A9 A0A1B0G9X9 A0A182F2W0 W5J7P6 E2AQ81 B0X5H6 A0A182NBS6 A0A026WWC1 A0A182R5I3 A0A182J5D0 A0A182KU05 A0A182KGN3 Q16S73 A0A151XIU1 A0A154PBJ8 A0A182Y0Q0 A0A195FMW4 A0A182MDI6 A0A182HRW0 E9IDG2 A0A2A3EDC6 A0A1S4FRB9 A0A1Q3FQ46 Q6TLC7 A0A1Q3FQ74 A0A084WTZ6 A0A182GV19 Q6TLC6
Pubmed
10436937
26354079
28756777
7733681
22118469
18362917
+ More
19820115 20566863 28004739 20075255 20798317 25348373 25830018 24508170 30249741 17994087 21719571 21347285 24495485 18057021 27129103 25474469 24845553 25401762 26823975 26108605 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 16507169 20920257 23761445 20966253 17510324 25244985 21282665 24438588 26483478
19820115 20566863 28004739 20075255 20798317 25348373 25830018 24508170 30249741 17994087 21719571 21347285 24495485 18057021 27129103 25474469 24845553 25401762 26823975 26108605 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 29403074 16507169 20920257 23761445 20966253 17510324 25244985 21282665 24438588 26483478
EMBL
AF141930
AAF70819.1
KQ458842
KPJ04909.1
KZ150011
PZC75068.1
+ More
NWSH01001510 PCG71022.1 AB185110 BAT57339.1 KP657653 RSAL01000078 AKL78878.1 RVE48739.1 L40373 AAA92342.1 AGBW02009801 OWR49896.1 UFQS01002407 UFQT01002407 SSX13976.1 SSX33395.1 UFQT01001356 SSX30192.1 KQ971342 EFA03563.1 GFDL01005455 JAV29590.1 DS235005 EEB10310.1 GEZM01057053 JAV72475.1 AAZX01000612 GL448902 EFN83605.1 GDHF01031548 JAI20766.1 GAKP01002574 JAC56378.1 GBYB01011322 GBYB01011324 GBYB01011325 JAG81089.1 JAG81091.1 JAG81092.1 JXJN01023285 GBXI01012167 JAD02125.1 KK107139 QOIP01000003 EZA57651.1 RLU25181.1 CH479221 EDW34889.1 GL888153 EGI66450.1 KQ435694 KOX80903.1 ADTU01022394 KQ982482 KYQ55859.1 KQ977643 KYN00859.1 KZ288253 PBC30921.1 CH964272 EDW83598.1 KQ979608 KYN20268.1 GDHF01022405 JAI29909.1 KQ414637 KOC66997.1 GL436654 EFN71668.1 OUUW01000017 SPP88994.1 KQ981836 KYN35044.1 ACPB03011743 ACPB03011744 GAMC01002436 GAMC01002435 GAMC01002434 JAC04122.1 CH940647 EDW70247.1 KRF84807.1 GDKW01002347 JAI54248.1 GBBI01003743 JAC14969.1 CH902636 EDV44766.2 LBMM01000654 KMQ97868.1 KK852908 KDR13981.1 CH933809 EDW19473.1 CH916366 EDV97214.1 GBHO01035499 GBHO01035498 GBHO01035497 GBRD01017745 GBRD01017744 GDHC01021994 GDHC01013787 JAG08105.1 JAG08106.1 JAG08107.1 JAG48082.1 JAP96634.1 JAQ04842.1 JRES01000361 KNC31951.1 CH480845 EDW50919.1 CH954184 EDV45235.1 CM002916 KMZ07265.1 CM000161 EDW99324.1 GECZ01013560 GECZ01002644 JAS56209.1 JAS67125.1 GECU01036643 GECU01005354 JAS71063.1 JAT02353.1 AE014135 BT010000 AAF59327.4 AAQ22469.1 NEVH01007816 PNF35570.1 GEDC01027143 JAS10155.1 BT011359 AAR96151.1 KQ760672 OAD59439.1 PYGN01000418 PSN46724.1 KQ459890 KPJ19591.1 GEBQ01023249 JAT16728.1 GEBQ01015110 JAT24867.1 CVRI01000074 CRL08262.1 DQ378286 ABD64799.1 ODYU01012221 SOQ58414.1 CCAG010022213 ADMH02002010 ETN59996.1 GL441701 EFN64415.1 DS232379 EDS40861.1 KK107078 QOIP01000011 EZA60347.1 RLU16916.1 CH477681 EAT37303.1 KQ982080 KYQ60261.1 KQ434869 KZC09232.1 KQ981490 KYN41259.1 AXCM01004029 APCN01001766 GL762454 EFZ21399.1 KZ288293 PBC29021.1 GFDL01005413 JAV29632.1 AY395071 AAQ96727.1 GFDL01005409 JAV29636.1 ATLV01026970 KE525421 KFB53690.1 JXUM01018714 KQ560520 KXJ82007.1 AY395072 AAQ96728.1
NWSH01001510 PCG71022.1 AB185110 BAT57339.1 KP657653 RSAL01000078 AKL78878.1 RVE48739.1 L40373 AAA92342.1 AGBW02009801 OWR49896.1 UFQS01002407 UFQT01002407 SSX13976.1 SSX33395.1 UFQT01001356 SSX30192.1 KQ971342 EFA03563.1 GFDL01005455 JAV29590.1 DS235005 EEB10310.1 GEZM01057053 JAV72475.1 AAZX01000612 GL448902 EFN83605.1 GDHF01031548 JAI20766.1 GAKP01002574 JAC56378.1 GBYB01011322 GBYB01011324 GBYB01011325 JAG81089.1 JAG81091.1 JAG81092.1 JXJN01023285 GBXI01012167 JAD02125.1 KK107139 QOIP01000003 EZA57651.1 RLU25181.1 CH479221 EDW34889.1 GL888153 EGI66450.1 KQ435694 KOX80903.1 ADTU01022394 KQ982482 KYQ55859.1 KQ977643 KYN00859.1 KZ288253 PBC30921.1 CH964272 EDW83598.1 KQ979608 KYN20268.1 GDHF01022405 JAI29909.1 KQ414637 KOC66997.1 GL436654 EFN71668.1 OUUW01000017 SPP88994.1 KQ981836 KYN35044.1 ACPB03011743 ACPB03011744 GAMC01002436 GAMC01002435 GAMC01002434 JAC04122.1 CH940647 EDW70247.1 KRF84807.1 GDKW01002347 JAI54248.1 GBBI01003743 JAC14969.1 CH902636 EDV44766.2 LBMM01000654 KMQ97868.1 KK852908 KDR13981.1 CH933809 EDW19473.1 CH916366 EDV97214.1 GBHO01035499 GBHO01035498 GBHO01035497 GBRD01017745 GBRD01017744 GDHC01021994 GDHC01013787 JAG08105.1 JAG08106.1 JAG08107.1 JAG48082.1 JAP96634.1 JAQ04842.1 JRES01000361 KNC31951.1 CH480845 EDW50919.1 CH954184 EDV45235.1 CM002916 KMZ07265.1 CM000161 EDW99324.1 GECZ01013560 GECZ01002644 JAS56209.1 JAS67125.1 GECU01036643 GECU01005354 JAS71063.1 JAT02353.1 AE014135 BT010000 AAF59327.4 AAQ22469.1 NEVH01007816 PNF35570.1 GEDC01027143 JAS10155.1 BT011359 AAR96151.1 KQ760672 OAD59439.1 PYGN01000418 PSN46724.1 KQ459890 KPJ19591.1 GEBQ01023249 JAT16728.1 GEBQ01015110 JAT24867.1 CVRI01000074 CRL08262.1 DQ378286 ABD64799.1 ODYU01012221 SOQ58414.1 CCAG010022213 ADMH02002010 ETN59996.1 GL441701 EFN64415.1 DS232379 EDS40861.1 KK107078 QOIP01000011 EZA60347.1 RLU16916.1 CH477681 EAT37303.1 KQ982080 KYQ60261.1 KQ434869 KZC09232.1 KQ981490 KYN41259.1 AXCM01004029 APCN01001766 GL762454 EFZ21399.1 KZ288293 PBC29021.1 GFDL01005413 JAV29632.1 AY395071 AAQ96727.1 GFDL01005409 JAV29636.1 ATLV01026970 KE525421 KFB53690.1 JXUM01018714 KQ560520 KXJ82007.1 AY395072 AAQ96728.1
Proteomes
UP000053268
UP000218220
UP000283053
UP000007151
UP000007266
UP000009046
+ More
UP000002358 UP000091820 UP000008237 UP000092460 UP000092443 UP000053097 UP000279307 UP000008744 UP000007755 UP000053105 UP000005205 UP000005203 UP000075809 UP000078542 UP000242457 UP000007798 UP000192221 UP000078492 UP000053825 UP000092445 UP000000311 UP000268350 UP000078541 UP000015103 UP000008792 UP000007801 UP000036403 UP000027135 UP000009192 UP000001070 UP000037069 UP000001292 UP000008711 UP000002282 UP000000803 UP000235965 UP000245037 UP000053240 UP000183832 UP000078200 UP000092444 UP000069272 UP000000673 UP000002320 UP000075884 UP000075900 UP000075880 UP000075882 UP000075881 UP000008820 UP000076502 UP000076408 UP000075883 UP000075840 UP000030765 UP000069940 UP000249989
UP000002358 UP000091820 UP000008237 UP000092460 UP000092443 UP000053097 UP000279307 UP000008744 UP000007755 UP000053105 UP000005205 UP000005203 UP000075809 UP000078542 UP000242457 UP000007798 UP000192221 UP000078492 UP000053825 UP000092445 UP000000311 UP000268350 UP000078541 UP000015103 UP000008792 UP000007801 UP000036403 UP000027135 UP000009192 UP000001070 UP000037069 UP000001292 UP000008711 UP000002282 UP000000803 UP000235965 UP000245037 UP000053240 UP000183832 UP000078200 UP000092444 UP000069272 UP000000673 UP000002320 UP000075884 UP000075900 UP000075880 UP000075882 UP000075881 UP000008820 UP000076502 UP000076408 UP000075883 UP000075840 UP000030765 UP000069940 UP000249989
Interpro
Gene 3D
ProteinModelPortal
Q9NJI0
A0A0N0PA64
A0A2W1BJM4
A0A2A4JHY8
A0A0U4ZU62
A0A0G3VIB4
+ More
Q25512 A0A212F833 A0A336L7S6 A0A336MIE5 D6WKL1 A0A1Q3FPR1 E0VAA4 A0A1Y1LKL2 K7IX81 A0A1A9WQD1 E2BL27 A0A0K8U2S1 A0A034WRI5 A0A0C9RDY6 A0A1B0BZS7 A0A1A9YA80 A0A0A1WUC9 A0A026WPT1 B4H889 F4WH84 A0A0M9AD94 A0A158NPD8 A0A088AT93 A0A151X6C7 A0A151IH15 A0A2A3EIS8 B4NHM9 A0A1W4V4Y7 A0A195E530 A0A0K8UT91 A0A0L7R7Y6 A0A1A9ZJY6 E2A4G9 A0A3B0K464 A0A195F4N9 T1I4T4 W8C964 B4LF59 A0A0N7Z8U8 A0A023F218 A0A1W4VH63 B3MZY1 A0A0J7L5P3 A0A067R6S2 B4KVK2 B4J3B5 A0A0A9WTG2 A0A0L0CI82 B4IIX8 B3P9S2 A0A0J9S2H0 B4PW19 A0A1B6GXH5 A0A1B6JT61 Q9V4E7 A0A2J7R419 A0A1B6CA44 Q6NND1 A0A310SS41 A0A2P8YR78 A0A0N1IJP4 A0A1B6KZ53 A0A1B6LMH6 A0A1J1J9R4 Q20C86 A0A1A9VFK0 A0A2H1X1A9 A0A1B0G9X9 A0A182F2W0 W5J7P6 E2AQ81 B0X5H6 A0A182NBS6 A0A026WWC1 A0A182R5I3 A0A182J5D0 A0A182KU05 A0A182KGN3 Q16S73 A0A151XIU1 A0A154PBJ8 A0A182Y0Q0 A0A195FMW4 A0A182MDI6 A0A182HRW0 E9IDG2 A0A2A3EDC6 A0A1S4FRB9 A0A1Q3FQ46 Q6TLC7 A0A1Q3FQ74 A0A084WTZ6 A0A182GV19 Q6TLC6
Q25512 A0A212F833 A0A336L7S6 A0A336MIE5 D6WKL1 A0A1Q3FPR1 E0VAA4 A0A1Y1LKL2 K7IX81 A0A1A9WQD1 E2BL27 A0A0K8U2S1 A0A034WRI5 A0A0C9RDY6 A0A1B0BZS7 A0A1A9YA80 A0A0A1WUC9 A0A026WPT1 B4H889 F4WH84 A0A0M9AD94 A0A158NPD8 A0A088AT93 A0A151X6C7 A0A151IH15 A0A2A3EIS8 B4NHM9 A0A1W4V4Y7 A0A195E530 A0A0K8UT91 A0A0L7R7Y6 A0A1A9ZJY6 E2A4G9 A0A3B0K464 A0A195F4N9 T1I4T4 W8C964 B4LF59 A0A0N7Z8U8 A0A023F218 A0A1W4VH63 B3MZY1 A0A0J7L5P3 A0A067R6S2 B4KVK2 B4J3B5 A0A0A9WTG2 A0A0L0CI82 B4IIX8 B3P9S2 A0A0J9S2H0 B4PW19 A0A1B6GXH5 A0A1B6JT61 Q9V4E7 A0A2J7R419 A0A1B6CA44 Q6NND1 A0A310SS41 A0A2P8YR78 A0A0N1IJP4 A0A1B6KZ53 A0A1B6LMH6 A0A1J1J9R4 Q20C86 A0A1A9VFK0 A0A2H1X1A9 A0A1B0G9X9 A0A182F2W0 W5J7P6 E2AQ81 B0X5H6 A0A182NBS6 A0A026WWC1 A0A182R5I3 A0A182J5D0 A0A182KU05 A0A182KGN3 Q16S73 A0A151XIU1 A0A154PBJ8 A0A182Y0Q0 A0A195FMW4 A0A182MDI6 A0A182HRW0 E9IDG2 A0A2A3EDC6 A0A1S4FRB9 A0A1Q3FQ46 Q6TLC7 A0A1Q3FQ74 A0A084WTZ6 A0A182GV19 Q6TLC6
PDB
4XPF
E-value=4.32097e-139,
Score=1269
Ontologies
GO
PANTHER
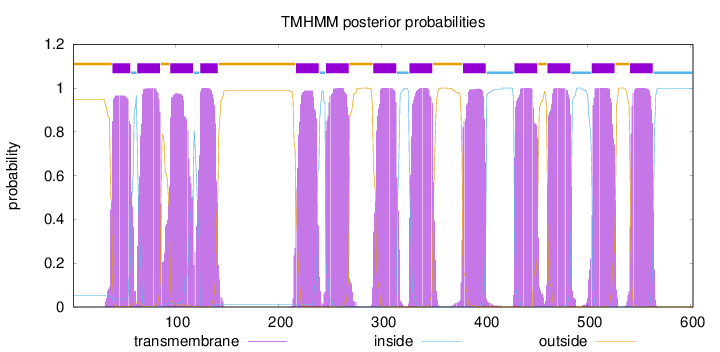
Topology
Length:
602
Number of predicted TMHs:
13
Exp number of AAs in TMHs:
284.34673
Exp number, first 60 AAs:
19.61672
Total prob of N-in:
0.05249
POSSIBLE N-term signal
sequence
outside
1 - 38
TMhelix
39 - 56
inside
57 - 62
TMhelix
63 - 85
outside
86 - 94
TMhelix
95 - 117
inside
118 - 123
TMhelix
124 - 141
outside
142 - 216
TMhelix
217 - 239
inside
240 - 245
TMhelix
246 - 268
outside
269 - 291
TMhelix
292 - 314
inside
315 - 326
TMhelix
327 - 349
outside
350 - 378
TMhelix
379 - 401
inside
402 - 428
TMhelix
429 - 451
outside
452 - 460
TMhelix
461 - 483
inside
484 - 503
TMhelix
504 - 526
outside
527 - 540
TMhelix
541 - 563
inside
564 - 602
Population Genetic Test Statistics
Pi
324.076623
Theta
170.416225
Tajima's D
2.907882
CLR
0.30676
CSRT
0.973801309934503
Interpretation
Uncertain
