Gene
KWMTBOMO16065
Pre Gene Modal
BGIBMGA004579
Annotation
PREDICTED:_calcium-dependent_secretion_activator_isoform_X9_[Bombyx_mori]
Full name
Calcium-dependent secretion activator
Alternative Name
Calcium-activated protein for secretion
Location in the cell
Cytoplasmic Reliability : 3.144
Sequence
CDS
ATGCCGAAGTTTGTGCTTAAAGAAATGGAATCCCTGTACATTGAGGAGCTGAAGTCATCCATCAACCTCCTCATGGCGAATCTGGAGTCCCTGCCCGTGTCGAAGGGAGGGGCGGACTCCAAATACGGACTCCACAAAATCAAGCGGTATAACCACAGATCCCAAGGCTCTTTAGCCAACAAACTGACGGGAGAAACGGACGGCGAAGTTGACACGCAGCTCACCAAAATGGACGTTGTGCTAACATTCCAAATCGAGGTAGTCGTCATGGAGGTTAAGGGTCTGAAGTCATTGGCACCAAATAGAATAGTTTACTGCACTATGGAAGTGGAGGGTGGTGATAAACTGCAGACTGATCAAGCTGAGGCGTCCAAGCCTATGTGGGACACGCAAGGTGATTTTAGTACAACGCAGCCTCTTCCAGCCGTGAAAGTGAAACTTTACACAGAGAACCCCGGAGTTTTGGCTTTGGAAGACAAAGAACTGGGGAAAGTGGTCTTAAGACCTACACCGTTGTCTAGTAAGGCGCCGGAGTGGCACAGGATGACGGTTCCGAAGAATCTACCTGACCAGGACTTGAGGATCAAGATCGCGTGTCGGATGGACAAGCCTTTGAATATGAAGCATTGCGGCTACTTGCACGTAATAGGGAAGAACGTGTGGCGGAAATGGAAACGCCGGTACATGGTGCTCGTGCAGGTCAGCCAGTACACCTTCGCGCTTTGCTCCTATAAAGACAAGAAGTCAGAGCCGGCCGAGATGATGCAGCTCGACGGCTTCACTGTCGACTACATAGAGCTCGCAAGCGCCCAACTCATCGTTGGACAAGGTAAGAAAGAAAATTTCAACACTAAACTGGAAGGCGCGAAGTACTTCATGAACGCGGTCCGCGACGGAGAGTCGGTGCTGATGGGAACCACAGACGAGAACGAGTGCCATCTGTGGGTGATGGCGTTGTACAGAGCGACGGGACAAAGTCACAAACCCACTCCCCCCACCACGTCCGATACGCATCACAAGATAATGGGAGACGCAGACAAAGCTCGTAAGCACGGAATGGAGGACTACATCCAGGCGGACCCGTGCCAGTTCGACCACCACCAACTGTTCTGCACGCTGCAGTCGCTGACCTTGAAATATCGACTACAAGACCCTTATTGCTCGCTGGGTTGGTTCTCCCCCGGCCAAGTGTTCGTCCTGGACGAGTACTGCGCCCGGTACGGAGTGCGAGGCTGCTATCGACACCTCTGCTACCTCTCCGACCTTTTGGACGTGGCGGAGAGCGGGCAGCAGACCGTGGACCCGACCCTCATGCACTACTCCTTCGCGTTCTGCGCCAGCCACGTGCACGGGAACAGTACCGCGACGTTGCCTGGCAGGCCAGACGGCGTTGGCAGCATCACCGTCCAGGAGAAGGAGAAGTTCGCCGAGATCAAAGAGCGGCTGAAGACGTTACTGCAACACCAAATAACTAACTTTAGATACGCATTCCCGTTCGGAAGACCGGAGGGAGCTCTGAAGGCTACGCTGTCGCTCTTGGAGAGAGTGCTGATGAAAGACGTGGTGACACCGGTGGCGCCGGAGGAGGTGCGGACAATGATACAGACGAGCTTGGAGAACGCGGCGCTTCTGAACTACACGCAGCTCAGTCAGAAAGCCAACATCGAAGATCTCAGAGGCGATACCATGGTAACGCCAGCTAAAAAATTAGAAGATCTCATACATCTGGCAGAACTCTGCGTTGACCTACTACAGCAAAACGAAGAACATTACGCGGAGGTTTATAACACAAAACCGGATTCTAGCGGTCAACAAAATGCATTTGCATGGTTCTCTGAACTTCTGGTGGAACACGCTGAGATCTTCTGGGCACTGTTCGCCGTGGACATGGACCGCGTTCTGTCCGAGCAGCCACCGGACACATGGGATTCGTTCCCACTCTTCCAAATATTAAACGACTATTTAAGAACAGATACTGATACCGAAGGAACATACCCAATGGATAATTTAAAAGGTGGCAGATTCCACGAACATCTCCGAGATACGTTTGCGCCATTGGTTGTACGTTACGTGGACCTGATGGAGAGTTCTATAGCTCAATCACTTCATAAAGGTTTCGAAAAGGAGCGATGGGAAATAAAGGGTAATGGCTGCGCGACTAGCGAAGAGTTGTTCTGGAAGCTGGACGCGTTGCAGTGCTTCATCAGAGACCTTCACTGGCCAGAGCCGGAGTTCCGTTCACACCTTGAACAGCGACTCAAACTCATGGCTAGTGACATGATGGAAACTTTAATACAGAGGACCGAGGCTGCTTTTCAGTCGTGGTTGAAGAAGAGCGTCACTTTCATGTCCACGGACTACATACTGCCGGCCGAGACGTGCGCGATGGTCAACGTCGCGCTGGACGCGAAGAACCAGGCCCTCAAGCTCTGCGCAGTTGAAGGGGTGGATATTTATCAGTACCACGCCAAAATGGACGGTCAAATCGAGGCCTGTCTAGTGGCGATGGCAACTGGCATGACAACAAGACTGTCCGCTGTTCTCGAAGCGACCTTAGCTAAGATTGCAAGATTCGATGAGGGAAGCCTTATCGGCTCCATCCTTACAATTGCCAACGTGTCCGGCTCTGGCAAGGACATCGGTCAGGGCTACGTCAACTTCATGAGAAACTCCATGGACCAAATCAGATCAAAGGTAACCGACGAGCTCTGGATCCTGCAGCTCTTCGAGCAATGGTACGGGAACCAGGTGACGATGATCAACAATTGGTTGTCAGAACGACCGGCGTTACATCCTCACCAGGTCGCCTGCCTCTCCATCTTGTTAAAGAAAATGTACAGCGACTTCGAGCTACAGGGGGTCATTGATGACAAGTTAAACTCTAAGACGTACCAGGGAGTCGCGGCCCGGATGCACACTGAAGAAGCGACGTGCAGCCTGTTGCTGAGTCAGCAAGACGGCGGTGGCGGTGACGATGGTGGTTCCGAAGAAGGCTCAAGAGGTGGAAAGTCGAAACTAGAAGCCCTCACCGAGGAAGCTAAGCTAGGCAACGTTACGGCTGTAGTTGGAAAGGTTGGAAATATGTTTGGTCGCGGCATAGGAGAACTGTCAACGAAACTAGGAGGCGCTTCGTCATGGTTCTAG
Protein
MPKFVLKEMESLYIEELKSSINLLMANLESLPVSKGGADSKYGLHKIKRYNHRSQGSLANKLTGETDGEVDTQLTKMDVVLTFQIEVVVMEVKGLKSLAPNRIVYCTMEVEGGDKLQTDQAEASKPMWDTQGDFSTTQPLPAVKVKLYTENPGVLALEDKELGKVVLRPTPLSSKAPEWHRMTVPKNLPDQDLRIKIACRMDKPLNMKHCGYLHVIGKNVWRKWKRRYMVLVQVSQYTFALCSYKDKKSEPAEMMQLDGFTVDYIELASAQLIVGQGKKENFNTKLEGAKYFMNAVRDGESVLMGTTDENECHLWVMALYRATGQSHKPTPPTTSDTHHKIMGDADKARKHGMEDYIQADPCQFDHHQLFCTLQSLTLKYRLQDPYCSLGWFSPGQVFVLDEYCARYGVRGCYRHLCYLSDLLDVAESGQQTVDPTLMHYSFAFCASHVHGNSTATLPGRPDGVGSITVQEKEKFAEIKERLKTLLQHQITNFRYAFPFGRPEGALKATLSLLERVLMKDVVTPVAPEEVRTMIQTSLENAALLNYTQLSQKANIEDLRGDTMVTPAKKLEDLIHLAELCVDLLQQNEEHYAEVYNTKPDSSGQQNAFAWFSELLVEHAEIFWALFAVDMDRVLSEQPPDTWDSFPLFQILNDYLRTDTDTEGTYPMDNLKGGRFHEHLRDTFAPLVVRYVDLMESSIAQSLHKGFEKERWEIKGNGCATSEELFWKLDALQCFIRDLHWPEPEFRSHLEQRLKLMASDMMETLIQRTEAAFQSWLKKSVTFMSTDYILPAETCAMVNVALDAKNQALKLCAVEGVDIYQYHAKMDGQIEACLVAMATGMTTRLSAVLEATLAKIARFDEGSLIGSILTIANVSGSGKDIGQGYVNFMRNSMDQIRSKVTDELWILQLFEQWYGNQVTMINNWLSERPALHPHQVACLSILLKKMYSDFELQGVIDDKLNSKTYQGVAARMHTEEATCSLLLSQQDGGGGDDGGSEEGSRGGKSKLEALTEEAKLGNVTAVVGKVGNMFGRGIGELSTKLGGASSWF
Summary
Description
Calcium-binding protein involved in exocytosis of vesicles filled with neurotransmitters and neuropeptides. May specifically mediate the Ca(2+)-dependent exocytosis of large dense-core vesicles (DCVs) and other dense-core vesicles. However, it probably also participates in small clear synaptic vesicles (SVs) exocytosis and it is unclear whether its function is related to Ca(2+) triggering.
Keywords
Alternative splicing
Calcium
Cell junction
Complete proteome
Cytoplasmic vesicle
Exocytosis
Lipid-binding
Membrane
Metal-binding
Protein transport
Reference proteome
Synapse
Transport
Feature
chain Calcium-dependent secretion activator
splice variant In isoform 3.
splice variant In isoform 3.
Uniprot
H9J4Y9
A0A0N0PEE3
A0A2H1W8I2
A0A0N0PA63
A0A2A4JN00
A0A2A4JLM5
+ More
A0A2A4JNH4 A0A2A4JMA7 A0A2A4JME3 A0A2A4JNE7 A0A2A4JML3 A0A2A4JNG7 A0A2A4JLV3 A0A2A4JLK0 A0A1Y1NFK5 A0A1W4WR25 A0A1W4WF75 A0A0T6BCC2 A0A1Y1NKE4 A0A2J7QJ63 A0A023EWW7 A0A069DY17 A0A067R127 E0VZ67 Q9NHE5-7 A0A1W4WBT6 A0A0Q9WWP0 A0A0Q5UX39 A0A0K8V7Z7 A0A0K8U112 A0A0P8XJH8 A0A0Q9XNT8 E2AI42 A0A151IMN5 A0A3L8D724 A0A2J7QJ73 A0A026W1K1 A0A0L0C5Q7 W8B7Q0 A0A158NPM9 A0A195EL55 A0A195ESH6 F4W7M3 A0A1A9WFC3 A0A151WJ46 A0A1B0BQD5 A0A1B0G6G9 A0A1A9ZSL9 A0A1A9V6J9 A0A195AT40 A0A1A9YP30 A0A1I8PC61 A0A1I8MUB9 A0A336M007 A0A0R1E7M4 A0A336KFR6 A0A1D2MT12 D6W738 E9J9W3 A0A154PMK3 A0A0L7QL63 E2C3B1 A0A0N0BG10 V9IDM1 A0A088AUU4 A0A310SMY2 A0A0A9Y0M3 A0A0A9Y4Q5 E9FY77 A0A0P5BZT2 A0A0P5BZX1 A0A0P5ARW3 N6T9E0 A0A0P5CC19 A0A0P6JE70 A0A0P5C007 A0A0J7KKK1 A0A0P4YEE4 A0A0N8A6J0 A0A0P5XHU0 A0A0P5C194 A0A0P4ZZC5 A0A0P4WRQ3 A0A0P5DMU3 A0A034W024 A0A0P5DEF1 K7IXW6 A0A087TFQ3 A0A2A3E199 A0A164QUF7 A0A0P5ZJ57 A0A0P5ZQ14 A0A0P6ACZ0 A0A0P5JPQ8 A0A0P5C1A3 A0A0P6FH06 A0A0P6D3D7 A0A0P4YKS6 A0A0N8A6J7 A0A0P5AT02
A0A2A4JNH4 A0A2A4JMA7 A0A2A4JME3 A0A2A4JNE7 A0A2A4JML3 A0A2A4JNG7 A0A2A4JLV3 A0A2A4JLK0 A0A1Y1NFK5 A0A1W4WR25 A0A1W4WF75 A0A0T6BCC2 A0A1Y1NKE4 A0A2J7QJ63 A0A023EWW7 A0A069DY17 A0A067R127 E0VZ67 Q9NHE5-7 A0A1W4WBT6 A0A0Q9WWP0 A0A0Q5UX39 A0A0K8V7Z7 A0A0K8U112 A0A0P8XJH8 A0A0Q9XNT8 E2AI42 A0A151IMN5 A0A3L8D724 A0A2J7QJ73 A0A026W1K1 A0A0L0C5Q7 W8B7Q0 A0A158NPM9 A0A195EL55 A0A195ESH6 F4W7M3 A0A1A9WFC3 A0A151WJ46 A0A1B0BQD5 A0A1B0G6G9 A0A1A9ZSL9 A0A1A9V6J9 A0A195AT40 A0A1A9YP30 A0A1I8PC61 A0A1I8MUB9 A0A336M007 A0A0R1E7M4 A0A336KFR6 A0A1D2MT12 D6W738 E9J9W3 A0A154PMK3 A0A0L7QL63 E2C3B1 A0A0N0BG10 V9IDM1 A0A088AUU4 A0A310SMY2 A0A0A9Y0M3 A0A0A9Y4Q5 E9FY77 A0A0P5BZT2 A0A0P5BZX1 A0A0P5ARW3 N6T9E0 A0A0P5CC19 A0A0P6JE70 A0A0P5C007 A0A0J7KKK1 A0A0P4YEE4 A0A0N8A6J0 A0A0P5XHU0 A0A0P5C194 A0A0P4ZZC5 A0A0P4WRQ3 A0A0P5DMU3 A0A034W024 A0A0P5DEF1 K7IXW6 A0A087TFQ3 A0A2A3E199 A0A164QUF7 A0A0P5ZJ57 A0A0P5ZQ14 A0A0P6ACZ0 A0A0P5JPQ8 A0A0P5C1A3 A0A0P6FH06 A0A0P6D3D7 A0A0P4YKS6 A0A0N8A6J7 A0A0P5AT02
Pubmed
EMBL
BABH01025689
KQ459890
KPJ19586.1
ODYU01007014
SOQ49378.1
KQ458842
+ More
KPJ04906.1 NWSH01001042 PCG72964.1 PCG72957.1 PCG72960.1 PCG72959.1 PCG72956.1 PCG72963.1 PCG72958.1 PCG72962.1 PCG72961.1 PCG72955.1 GEZM01003808 JAV96549.1 LJIG01001968 KRT84972.1 GEZM01003803 JAV96556.1 NEVH01013560 PNF28627.1 GBBI01005090 JAC13622.1 GBGD01000109 JAC88780.1 KK852819 KDR15617.1 DS235849 EEB18673.1 AF223578 AE014135 AY118427 AF145637 CH940665 KRF85291.1 CH954250 KQS47970.1 GDHF01017318 JAI34996.1 GDHF01032299 JAI20015.1 CH902646 KPU74920.1 CH933813 KRG07338.1 GL439639 EFN66893.1 KQ977026 KYN06215.1 QOIP01000012 RLU16079.1 PNF28630.1 KK107488 EZA49950.1 JRES01000956 KNC26759.1 GAMC01009315 JAB97240.1 ADTU01022623 ADTU01022624 ADTU01022625 KQ978730 KYN28990.1 KQ981986 KYN31200.1 GL887844 EGI69896.1 KQ983049 KYQ47856.1 JXJN01018561 CCAG010007966 KQ976745 KYM75388.1 UFQS01000373 UFQT01000373 SSX03300.1 SSX23666.1 CM000161 KRK05017.1 SSX03301.1 SSX23667.1 LJIJ01000606 ODM95924.1 KQ971307 EFA11414.2 GL769474 EFZ10391.1 KQ434954 KZC12528.1 KQ414934 KOC59352.1 GL452318 EFN77557.1 KQ435794 KOX74118.1 JR039550 AEY58752.1 KQ759866 OAD62492.1 GBHO01017012 JAG26592.1 GBHO01017013 JAG26591.1 GL732527 EFX87826.1 GDIP01177616 JAJ45786.1 GDIP01177562 JAJ45840.1 GDIP01209358 JAJ14044.1 APGK01047302 APGK01047303 APGK01047304 KB741077 KB631602 ENN74348.1 ERL84569.1 GDIP01177564 JAJ45838.1 GDIQ01018773 JAN75964.1 GDIP01182263 GDIP01145668 JAJ41139.1 LBMM01006277 KMQ90746.1 GDIP01229607 GDIP01195411 GDIP01180558 JAI93794.1 GDIP01177617 JAJ45785.1 GDIP01071875 JAM31840.1 GDIP01177565 JAJ45837.1 GDIP01210408 JAJ12994.1 GDIP01252552 GDIP01238561 GDIP01230707 GDIP01196458 GDIP01180559 GDIP01152365 GDIP01145667 GDIP01132067 GDIP01132066 GDIP01132065 GDIP01130866 JAI70849.1 GDIP01155115 JAJ68287.1 GAKP01010041 JAC48911.1 GDIP01162661 JAJ60741.1 KK115013 KFM63942.1 KZ288455 PBC25523.1 LRGB01002371 KZS08069.1 GDIP01044334 JAM59381.1 GDIP01044332 JAM59383.1 GDIP01044333 JAM59382.1 GDIQ01196984 JAK54741.1 GDIP01177566 JAJ45836.1 GDIQ01050245 JAN44492.1 GDIQ01083379 JAN11358.1 GDIP01229608 GDIP01219906 GDIP01071876 JAI93793.1 GDIP01177561 JAJ45841.1 GDIP01198410 JAJ24992.1
KPJ04906.1 NWSH01001042 PCG72964.1 PCG72957.1 PCG72960.1 PCG72959.1 PCG72956.1 PCG72963.1 PCG72958.1 PCG72962.1 PCG72961.1 PCG72955.1 GEZM01003808 JAV96549.1 LJIG01001968 KRT84972.1 GEZM01003803 JAV96556.1 NEVH01013560 PNF28627.1 GBBI01005090 JAC13622.1 GBGD01000109 JAC88780.1 KK852819 KDR15617.1 DS235849 EEB18673.1 AF223578 AE014135 AY118427 AF145637 CH940665 KRF85291.1 CH954250 KQS47970.1 GDHF01017318 JAI34996.1 GDHF01032299 JAI20015.1 CH902646 KPU74920.1 CH933813 KRG07338.1 GL439639 EFN66893.1 KQ977026 KYN06215.1 QOIP01000012 RLU16079.1 PNF28630.1 KK107488 EZA49950.1 JRES01000956 KNC26759.1 GAMC01009315 JAB97240.1 ADTU01022623 ADTU01022624 ADTU01022625 KQ978730 KYN28990.1 KQ981986 KYN31200.1 GL887844 EGI69896.1 KQ983049 KYQ47856.1 JXJN01018561 CCAG010007966 KQ976745 KYM75388.1 UFQS01000373 UFQT01000373 SSX03300.1 SSX23666.1 CM000161 KRK05017.1 SSX03301.1 SSX23667.1 LJIJ01000606 ODM95924.1 KQ971307 EFA11414.2 GL769474 EFZ10391.1 KQ434954 KZC12528.1 KQ414934 KOC59352.1 GL452318 EFN77557.1 KQ435794 KOX74118.1 JR039550 AEY58752.1 KQ759866 OAD62492.1 GBHO01017012 JAG26592.1 GBHO01017013 JAG26591.1 GL732527 EFX87826.1 GDIP01177616 JAJ45786.1 GDIP01177562 JAJ45840.1 GDIP01209358 JAJ14044.1 APGK01047302 APGK01047303 APGK01047304 KB741077 KB631602 ENN74348.1 ERL84569.1 GDIP01177564 JAJ45838.1 GDIQ01018773 JAN75964.1 GDIP01182263 GDIP01145668 JAJ41139.1 LBMM01006277 KMQ90746.1 GDIP01229607 GDIP01195411 GDIP01180558 JAI93794.1 GDIP01177617 JAJ45785.1 GDIP01071875 JAM31840.1 GDIP01177565 JAJ45837.1 GDIP01210408 JAJ12994.1 GDIP01252552 GDIP01238561 GDIP01230707 GDIP01196458 GDIP01180559 GDIP01152365 GDIP01145667 GDIP01132067 GDIP01132066 GDIP01132065 GDIP01130866 JAI70849.1 GDIP01155115 JAJ68287.1 GAKP01010041 JAC48911.1 GDIP01162661 JAJ60741.1 KK115013 KFM63942.1 KZ288455 PBC25523.1 LRGB01002371 KZS08069.1 GDIP01044334 JAM59381.1 GDIP01044332 JAM59383.1 GDIP01044333 JAM59382.1 GDIQ01196984 JAK54741.1 GDIP01177566 JAJ45836.1 GDIQ01050245 JAN44492.1 GDIQ01083379 JAN11358.1 GDIP01229608 GDIP01219906 GDIP01071876 JAI93793.1 GDIP01177561 JAJ45841.1 GDIP01198410 JAJ24992.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000192223
UP000235965
+ More
UP000027135 UP000009046 UP000000803 UP000192221 UP000008792 UP000008711 UP000007801 UP000009192 UP000000311 UP000078542 UP000279307 UP000053097 UP000037069 UP000005205 UP000078492 UP000078541 UP000007755 UP000091820 UP000075809 UP000092460 UP000092444 UP000092445 UP000078200 UP000078540 UP000092443 UP000095300 UP000095301 UP000002282 UP000094527 UP000007266 UP000076502 UP000053825 UP000008237 UP000053105 UP000005203 UP000000305 UP000019118 UP000030742 UP000036403 UP000002358 UP000054359 UP000242457 UP000076858
UP000027135 UP000009046 UP000000803 UP000192221 UP000008792 UP000008711 UP000007801 UP000009192 UP000000311 UP000078542 UP000279307 UP000053097 UP000037069 UP000005205 UP000078492 UP000078541 UP000007755 UP000091820 UP000075809 UP000092460 UP000092444 UP000092445 UP000078200 UP000078540 UP000092443 UP000095300 UP000095301 UP000002282 UP000094527 UP000007266 UP000076502 UP000053825 UP000008237 UP000053105 UP000005203 UP000000305 UP000019118 UP000030742 UP000036403 UP000002358 UP000054359 UP000242457 UP000076858
Interpro
IPR001849
PH_domain
+ More
IPR011993 PH-like_dom_sf
IPR014770 Munc13_1
IPR033227 CAPS
IPR010439 CAPS_dom
IPR000008 C2_dom
IPR035892 C2_domain_sf
IPR017946 PLC-like_Pdiesterase_TIM-brl
IPR000909 PLipase_C_PInositol-sp_X_dom
IPR011992 EF-hand-dom_pair
IPR001192 PI-PLC_fam
IPR001711 PLipase_C_Pinositol-sp_Y
IPR011993 PH-like_dom_sf
IPR014770 Munc13_1
IPR033227 CAPS
IPR010439 CAPS_dom
IPR000008 C2_dom
IPR035892 C2_domain_sf
IPR017946 PLC-like_Pdiesterase_TIM-brl
IPR000909 PLipase_C_PInositol-sp_X_dom
IPR011992 EF-hand-dom_pair
IPR001192 PI-PLC_fam
IPR001711 PLipase_C_Pinositol-sp_Y
Gene 3D
ProteinModelPortal
H9J4Y9
A0A0N0PEE3
A0A2H1W8I2
A0A0N0PA63
A0A2A4JN00
A0A2A4JLM5
+ More
A0A2A4JNH4 A0A2A4JMA7 A0A2A4JME3 A0A2A4JNE7 A0A2A4JML3 A0A2A4JNG7 A0A2A4JLV3 A0A2A4JLK0 A0A1Y1NFK5 A0A1W4WR25 A0A1W4WF75 A0A0T6BCC2 A0A1Y1NKE4 A0A2J7QJ63 A0A023EWW7 A0A069DY17 A0A067R127 E0VZ67 Q9NHE5-7 A0A1W4WBT6 A0A0Q9WWP0 A0A0Q5UX39 A0A0K8V7Z7 A0A0K8U112 A0A0P8XJH8 A0A0Q9XNT8 E2AI42 A0A151IMN5 A0A3L8D724 A0A2J7QJ73 A0A026W1K1 A0A0L0C5Q7 W8B7Q0 A0A158NPM9 A0A195EL55 A0A195ESH6 F4W7M3 A0A1A9WFC3 A0A151WJ46 A0A1B0BQD5 A0A1B0G6G9 A0A1A9ZSL9 A0A1A9V6J9 A0A195AT40 A0A1A9YP30 A0A1I8PC61 A0A1I8MUB9 A0A336M007 A0A0R1E7M4 A0A336KFR6 A0A1D2MT12 D6W738 E9J9W3 A0A154PMK3 A0A0L7QL63 E2C3B1 A0A0N0BG10 V9IDM1 A0A088AUU4 A0A310SMY2 A0A0A9Y0M3 A0A0A9Y4Q5 E9FY77 A0A0P5BZT2 A0A0P5BZX1 A0A0P5ARW3 N6T9E0 A0A0P5CC19 A0A0P6JE70 A0A0P5C007 A0A0J7KKK1 A0A0P4YEE4 A0A0N8A6J0 A0A0P5XHU0 A0A0P5C194 A0A0P4ZZC5 A0A0P4WRQ3 A0A0P5DMU3 A0A034W024 A0A0P5DEF1 K7IXW6 A0A087TFQ3 A0A2A3E199 A0A164QUF7 A0A0P5ZJ57 A0A0P5ZQ14 A0A0P6ACZ0 A0A0P5JPQ8 A0A0P5C1A3 A0A0P6FH06 A0A0P6D3D7 A0A0P4YKS6 A0A0N8A6J7 A0A0P5AT02
A0A2A4JNH4 A0A2A4JMA7 A0A2A4JME3 A0A2A4JNE7 A0A2A4JML3 A0A2A4JNG7 A0A2A4JLV3 A0A2A4JLK0 A0A1Y1NFK5 A0A1W4WR25 A0A1W4WF75 A0A0T6BCC2 A0A1Y1NKE4 A0A2J7QJ63 A0A023EWW7 A0A069DY17 A0A067R127 E0VZ67 Q9NHE5-7 A0A1W4WBT6 A0A0Q9WWP0 A0A0Q5UX39 A0A0K8V7Z7 A0A0K8U112 A0A0P8XJH8 A0A0Q9XNT8 E2AI42 A0A151IMN5 A0A3L8D724 A0A2J7QJ73 A0A026W1K1 A0A0L0C5Q7 W8B7Q0 A0A158NPM9 A0A195EL55 A0A195ESH6 F4W7M3 A0A1A9WFC3 A0A151WJ46 A0A1B0BQD5 A0A1B0G6G9 A0A1A9ZSL9 A0A1A9V6J9 A0A195AT40 A0A1A9YP30 A0A1I8PC61 A0A1I8MUB9 A0A336M007 A0A0R1E7M4 A0A336KFR6 A0A1D2MT12 D6W738 E9J9W3 A0A154PMK3 A0A0L7QL63 E2C3B1 A0A0N0BG10 V9IDM1 A0A088AUU4 A0A310SMY2 A0A0A9Y0M3 A0A0A9Y4Q5 E9FY77 A0A0P5BZT2 A0A0P5BZX1 A0A0P5ARW3 N6T9E0 A0A0P5CC19 A0A0P6JE70 A0A0P5C007 A0A0J7KKK1 A0A0P4YEE4 A0A0N8A6J0 A0A0P5XHU0 A0A0P5C194 A0A0P4ZZC5 A0A0P4WRQ3 A0A0P5DMU3 A0A034W024 A0A0P5DEF1 K7IXW6 A0A087TFQ3 A0A2A3E199 A0A164QUF7 A0A0P5ZJ57 A0A0P5ZQ14 A0A0P6ACZ0 A0A0P5JPQ8 A0A0P5C1A3 A0A0P6FH06 A0A0P6D3D7 A0A0P4YKS6 A0A0N8A6J7 A0A0P5AT02
PDB
6A68
E-value=3.04298e-71,
Score=687
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasmic vesicle membrane
Cell junction
Synapse
Cell junction
Synapse
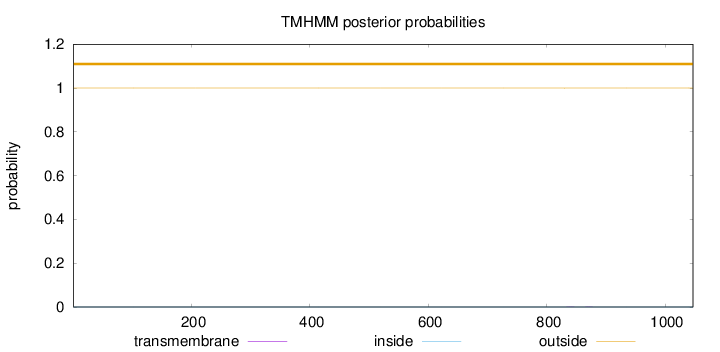
Length:
1047
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00742999999999999
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.00005
outside
1 - 1047
Population Genetic Test Statistics
Pi
283.936666
Theta
193.845282
Tajima's D
1.358901
CLR
0.108099
CSRT
0.756562171891405
Interpretation
Uncertain
