Gene
KWMTBOMO16064 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004580
Annotation
PREDICTED:_protein-methionine_sulfoxide_oxidase_mical3a_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.261
Sequence
CDS
ATGGCGCTAGCGACGGCCAAAGCGACGATGGAGTCCCAGAACCTCATCCAAACCGAGTCGAGGGTCCAGCACAGCTATCAGGAGGTGGTCACCGAGCAGCGCCAGATAAAGAAGAAGTCTAAATCCACTAGACGGCACAAAGATGAAGGATCAATTTCTGTGTCTAAAAGCTCTGAAAAGATTTACAAGAAAATCAAAGCCGCCTCGGACGGGGAATCGAACCCGAAGTGCGAGAAGTGCGCCCGTCCCGTCTACGCCATGGAGAGGATCAAGGCTGAGCGGAGAGTCTGGCACAAGGACTGCTTCCGATGTGTCCAGTGCAACAAACAACTCACAGTGGAGACTTACCAGAGCGACCACACGACCTTATACTGCAAGCCTCACTTCAAGCATCTGTTCGAACCCAAACCTGTCGAAGATGACTACGAGATTGATGCAGCACCGAAGAAACACCAAATGATAATTTGCGAGAGCAACCCGGTCGAACTGCCGCCAGATGTCGTTAGAGCGTCCGACAAACCGGACCTAGGGCTGGAGGAGCTGGCTTCCCTGGACGTGAAGTCCAGGTTCGAAGTGTTCGAGCGCGCCGCCAAGGGCAGCGACGAGCCTCCGCCCCTCGAGCACCGGGCCCCGCGCGAGAAGAGCACCGCCCTGCTCTCCAAGCTAGCCAAATTCAAGGCGAAGGGCATGGACATCGGTGTTTCGGACGAATACCTCAATGGGGTTCCCGTAGAACCCAGCTCCAGCGAACATGAAGATGAAGAAGACGAAGACTCGATCCTCAAGAACTCGTACAAGCACAGCTCGAAGGGCGAGCAGCCGGTGTCCTTCTGCAACATGAGCGAGATAGTCGGCAGGTTCGAGAGCGGCCAGCACTCCGCCTCCGAGAGACACCGCGAGAGGAAACAGGAGATCCAGAACATCAGGAGCAGACTGTTCATGGGCAAACAGGCCAAGATCAAGGAGATGTACGAACAATCGGTCATGCAAAGCGAACAGAGCGTTACGTCAGCCGACAAAATAGCCCGCGACTTGGAACTGGATAAAGAGAAAGCTCGGGCCATAAAGCAGCGCTTCGAGAACGGCGAGCTTTTCAACGACGAAAACCAACCGCCCAGGAACAGGGAGATCGACGACAAAGCTCTCTTCAATGAAGGTATCGGTAAGAAGTCTCGTTCTATATTCCTGGAGCTGGATGCGAACGCCAAGAGCACGGGCCCCGCGTCGCCGCCGCCGCACCAGCCGCCGCGCCGCAAGGAGATCGTTAGTCGCTTCATCCACTATATTTGA
Protein
MALATAKATMESQNLIQTESRVQHSYQEVVTEQRQIKKKSKSTRRHKDEGSISVSKSSEKIYKKIKAASDGESNPKCEKCARPVYAMERIKAERRVWHKDCFRCVQCNKQLTVETYQSDHTTLYCKPHFKHLFEPKPVEDDYEIDAAPKKHQMIICESNPVELPPDVVRASDKPDLGLEELASLDVKSRFEVFERAAKGSDEPPPLEHRAPREKSTALLSKLAKFKAKGMDIGVSDEYLNGVPVEPSSSEHEDEEDEDSILKNSYKHSSKGEQPVSFCNMSEIVGRFESGQHSASERHRERKQEIQNIRSRLFMGKQAKIKEMYEQSVMQSEQSVTSADKIARDLELDKEKARAIKQRFENGELFNDENQPPRNREIDDKALFNEGIGKKSRSIFLELDANAKSTGPASPPPHQPPRRKEIVSRFIHYI
Summary
Uniprot
A0A2A4JQ30
A0A2A4JQX5
A0A0N1IN34
A0A0N1IQ26
A0A212ES59
A0A1B6CU28
+ More
A0A1B6EGA4 A0A1Y1MFW3 A0A1Y1MCG1 A0A2S2Q6M0 A0A2S2QGV5 A0A2H8TNT3 A0A1J1IJL1 T1HJN0 A0A088AF55 A0A067RKX2 J9JQA5 K7IUC4 D6WYU2 E2A256 A0A0C9QNW9 A0A2J7PXS6 A0A0C9R6E0 A0A2J7PXQ6 A0A2J7PXR8 E2C1L4 A0A232FHR6 A0A154PH31 A0A2J7PXS3 A0A2J7PXS1 A0A158N9E7 A0A2J7PXR5 A0A2J7PXU4 A0A2J7PXT3 A0A2J7PXR9 A0A151ILQ7 A0A151IUM6 A0A195FH09 A0A151I1A4 A0A2J7PXT0 A0A2J7PXS8 A0A2J7PXQ5 A0A2J7PXS7 R4WPQ9 A0A2J7PXU1 A0A336L4I9 A0A0A1XRX3 A0A1L8E5U1 A0A1B6CMR6 A0A1B0CD21 A0A1B6DXF3 A0A310S6J9 A0A034VDC1 A0A034VB80 A0A034VCD3 A0A034VDZ5 A0A034VB88 N6U922 A0A1Y1MCG0 A0A1B6CP60 A0A1B6CPR2 A0A1B6C872 A0A1B6EBX8 A0A1B6C321 A0A1B6DWQ7 A0A1B6D707 A0A1Y1MCV8 A0A0K8TY27 A0A0K8WAU4 F4WEA4 A0A0Q9WUR7 A0A151WEM0 A0A069DWI6 A0A0M4F7E1 A0A0C9RW42 A0A0P4VKQ0 A0A2J7PXR4 A0A146LYW5 A0A0Q9WL53 D0Z7B4 B4MF13 A0A026X480 A0A0P8ZIN0 A0A0J7KUA9 A0A1A9WQC6 E0VLU0 A0A1B6H1P8 A0A0R3P0T0 A0A0L7QYK3 B4H9Q9 A0A1W4WBT9 A0A1W4W145 A0A3B0KUD6 A0A3B0KK07 A0A3B0KQL8
A0A1B6EGA4 A0A1Y1MFW3 A0A1Y1MCG1 A0A2S2Q6M0 A0A2S2QGV5 A0A2H8TNT3 A0A1J1IJL1 T1HJN0 A0A088AF55 A0A067RKX2 J9JQA5 K7IUC4 D6WYU2 E2A256 A0A0C9QNW9 A0A2J7PXS6 A0A0C9R6E0 A0A2J7PXQ6 A0A2J7PXR8 E2C1L4 A0A232FHR6 A0A154PH31 A0A2J7PXS3 A0A2J7PXS1 A0A158N9E7 A0A2J7PXR5 A0A2J7PXU4 A0A2J7PXT3 A0A2J7PXR9 A0A151ILQ7 A0A151IUM6 A0A195FH09 A0A151I1A4 A0A2J7PXT0 A0A2J7PXS8 A0A2J7PXQ5 A0A2J7PXS7 R4WPQ9 A0A2J7PXU1 A0A336L4I9 A0A0A1XRX3 A0A1L8E5U1 A0A1B6CMR6 A0A1B0CD21 A0A1B6DXF3 A0A310S6J9 A0A034VDC1 A0A034VB80 A0A034VCD3 A0A034VDZ5 A0A034VB88 N6U922 A0A1Y1MCG0 A0A1B6CP60 A0A1B6CPR2 A0A1B6C872 A0A1B6EBX8 A0A1B6C321 A0A1B6DWQ7 A0A1B6D707 A0A1Y1MCV8 A0A0K8TY27 A0A0K8WAU4 F4WEA4 A0A0Q9WUR7 A0A151WEM0 A0A069DWI6 A0A0M4F7E1 A0A0C9RW42 A0A0P4VKQ0 A0A2J7PXR4 A0A146LYW5 A0A0Q9WL53 D0Z7B4 B4MF13 A0A026X480 A0A0P8ZIN0 A0A0J7KUA9 A0A1A9WQC6 E0VLU0 A0A1B6H1P8 A0A0R3P0T0 A0A0L7QYK3 B4H9Q9 A0A1W4WBT9 A0A1W4W145 A0A3B0KUD6 A0A3B0KK07 A0A3B0KQL8
Pubmed
EMBL
NWSH01000793
PCG74175.1
PCG74176.1
KQ458842
KPJ04905.1
KQ459890
+ More
KPJ19585.1 AGBW02012867 OWR44317.1 GEDC01020391 JAS16907.1 GEDC01000326 JAS36972.1 GEZM01034927 GEZM01034926 JAV83520.1 GEZM01034941 GEZM01034928 JAV83519.1 GGMS01004048 MBY73251.1 GGMS01007765 MBY76968.1 GFXV01003776 MBW15581.1 CVRI01000048 CRK98633.1 ACPB03011838 ACPB03011839 ACPB03011840 ACPB03011841 ACPB03011842 ACPB03011843 ACPB03011844 KK852455 KDR23628.1 ABLF02027357 ABLF02027363 ABLF02050035 AAZX01003953 AAZX01006931 KQ971357 EFA08463.1 GL435922 EFN72461.1 GBYB01002322 JAG72089.1 NEVH01020852 PNF21135.1 GBYB01002321 JAG72088.1 PNF21128.1 PNF21127.1 GL451937 EFN78190.1 NNAY01000211 OXU29999.1 KQ434902 KZC11107.1 PNF21131.1 PNF21132.1 ADTU01000137 ADTU01000138 ADTU01000139 ADTU01000140 ADTU01000141 PNF21125.1 PNF21134.1 PNF21126.1 PNF21123.1 KQ977104 KYN05822.1 KQ980956 KYN11098.1 KQ981606 KYN39656.1 KQ976616 KYM79264.1 PNF21133.1 PNF21124.1 PNF21129.1 PNF21130.1 AK417651 BAN20866.1 PNF21136.1 UFQS01001714 UFQT01001714 SSX11938.1 SSX31485.1 GBXI01000565 JAD13727.1 GFDF01000070 JAV14014.1 GEDC01022576 JAS14722.1 AJWK01007325 GEDC01006945 JAS30353.1 KQ767877 OAD53257.1 GAKP01019197 JAC39755.1 GAKP01019203 JAC39749.1 GAKP01019195 JAC39757.1 GAKP01019194 JAC39758.1 GAKP01019193 JAC39759.1 APGK01044215 KB741021 ENN75067.1 GEZM01034922 JAV83522.1 GEDC01022144 JAS15154.1 GEDC01021953 JAS15345.1 GEDC01027664 JAS09634.1 GEDC01001901 JAS35397.1 GEDC01029693 JAS07605.1 GEDC01007229 JAS30069.1 GEDC01015827 JAS21471.1 GEZM01034943 JAV83513.1 GDHF01033133 JAI19181.1 GDHF01004102 JAI48212.1 GL888102 EGI67542.1 CH964272 KRF99803.1 KQ983238 KYQ46328.1 GBGD01000629 JAC88260.1 CP012527 ALC48100.1 GBYB01013165 JAG82932.1 GDKW01003511 JAI53084.1 PNF21122.1 GDHC01006803 JAQ11826.1 CH940665 KRF85370.1 CM000831 ACY70482.1 EDW71114.2 KK107026 EZA62184.1 CH902633 KPU74650.1 LBMM01003187 KMQ93839.1 DS235281 EEB14346.1 GECZ01001170 JAS68599.1 CH475446 KRT05279.1 KQ414688 KOC63684.1 CH479230 EDW36543.1 OUUW01000017 SPP88921.1 SPP88920.1 SPP88919.1
KPJ19585.1 AGBW02012867 OWR44317.1 GEDC01020391 JAS16907.1 GEDC01000326 JAS36972.1 GEZM01034927 GEZM01034926 JAV83520.1 GEZM01034941 GEZM01034928 JAV83519.1 GGMS01004048 MBY73251.1 GGMS01007765 MBY76968.1 GFXV01003776 MBW15581.1 CVRI01000048 CRK98633.1 ACPB03011838 ACPB03011839 ACPB03011840 ACPB03011841 ACPB03011842 ACPB03011843 ACPB03011844 KK852455 KDR23628.1 ABLF02027357 ABLF02027363 ABLF02050035 AAZX01003953 AAZX01006931 KQ971357 EFA08463.1 GL435922 EFN72461.1 GBYB01002322 JAG72089.1 NEVH01020852 PNF21135.1 GBYB01002321 JAG72088.1 PNF21128.1 PNF21127.1 GL451937 EFN78190.1 NNAY01000211 OXU29999.1 KQ434902 KZC11107.1 PNF21131.1 PNF21132.1 ADTU01000137 ADTU01000138 ADTU01000139 ADTU01000140 ADTU01000141 PNF21125.1 PNF21134.1 PNF21126.1 PNF21123.1 KQ977104 KYN05822.1 KQ980956 KYN11098.1 KQ981606 KYN39656.1 KQ976616 KYM79264.1 PNF21133.1 PNF21124.1 PNF21129.1 PNF21130.1 AK417651 BAN20866.1 PNF21136.1 UFQS01001714 UFQT01001714 SSX11938.1 SSX31485.1 GBXI01000565 JAD13727.1 GFDF01000070 JAV14014.1 GEDC01022576 JAS14722.1 AJWK01007325 GEDC01006945 JAS30353.1 KQ767877 OAD53257.1 GAKP01019197 JAC39755.1 GAKP01019203 JAC39749.1 GAKP01019195 JAC39757.1 GAKP01019194 JAC39758.1 GAKP01019193 JAC39759.1 APGK01044215 KB741021 ENN75067.1 GEZM01034922 JAV83522.1 GEDC01022144 JAS15154.1 GEDC01021953 JAS15345.1 GEDC01027664 JAS09634.1 GEDC01001901 JAS35397.1 GEDC01029693 JAS07605.1 GEDC01007229 JAS30069.1 GEDC01015827 JAS21471.1 GEZM01034943 JAV83513.1 GDHF01033133 JAI19181.1 GDHF01004102 JAI48212.1 GL888102 EGI67542.1 CH964272 KRF99803.1 KQ983238 KYQ46328.1 GBGD01000629 JAC88260.1 CP012527 ALC48100.1 GBYB01013165 JAG82932.1 GDKW01003511 JAI53084.1 PNF21122.1 GDHC01006803 JAQ11826.1 CH940665 KRF85370.1 CM000831 ACY70482.1 EDW71114.2 KK107026 EZA62184.1 CH902633 KPU74650.1 LBMM01003187 KMQ93839.1 DS235281 EEB14346.1 GECZ01001170 JAS68599.1 CH475446 KRT05279.1 KQ414688 KOC63684.1 CH479230 EDW36543.1 OUUW01000017 SPP88921.1 SPP88920.1 SPP88919.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000183832
UP000015103
+ More
UP000005203 UP000027135 UP000007819 UP000002358 UP000007266 UP000000311 UP000235965 UP000008237 UP000215335 UP000076502 UP000005205 UP000078542 UP000078492 UP000078541 UP000078540 UP000092461 UP000019118 UP000007755 UP000007798 UP000075809 UP000092553 UP000008792 UP000053097 UP000007801 UP000036403 UP000091820 UP000009046 UP000001819 UP000053825 UP000008744 UP000192221 UP000268350
UP000005203 UP000027135 UP000007819 UP000002358 UP000007266 UP000000311 UP000235965 UP000008237 UP000215335 UP000076502 UP000005205 UP000078542 UP000078492 UP000078541 UP000078540 UP000092461 UP000019118 UP000007755 UP000007798 UP000075809 UP000092553 UP000008792 UP000053097 UP000007801 UP000036403 UP000091820 UP000009046 UP000001819 UP000053825 UP000008744 UP000192221 UP000268350
Pfam
PF00412 LIM
Interpro
IPR001781
Znf_LIM
ProteinModelPortal
A0A2A4JQ30
A0A2A4JQX5
A0A0N1IN34
A0A0N1IQ26
A0A212ES59
A0A1B6CU28
+ More
A0A1B6EGA4 A0A1Y1MFW3 A0A1Y1MCG1 A0A2S2Q6M0 A0A2S2QGV5 A0A2H8TNT3 A0A1J1IJL1 T1HJN0 A0A088AF55 A0A067RKX2 J9JQA5 K7IUC4 D6WYU2 E2A256 A0A0C9QNW9 A0A2J7PXS6 A0A0C9R6E0 A0A2J7PXQ6 A0A2J7PXR8 E2C1L4 A0A232FHR6 A0A154PH31 A0A2J7PXS3 A0A2J7PXS1 A0A158N9E7 A0A2J7PXR5 A0A2J7PXU4 A0A2J7PXT3 A0A2J7PXR9 A0A151ILQ7 A0A151IUM6 A0A195FH09 A0A151I1A4 A0A2J7PXT0 A0A2J7PXS8 A0A2J7PXQ5 A0A2J7PXS7 R4WPQ9 A0A2J7PXU1 A0A336L4I9 A0A0A1XRX3 A0A1L8E5U1 A0A1B6CMR6 A0A1B0CD21 A0A1B6DXF3 A0A310S6J9 A0A034VDC1 A0A034VB80 A0A034VCD3 A0A034VDZ5 A0A034VB88 N6U922 A0A1Y1MCG0 A0A1B6CP60 A0A1B6CPR2 A0A1B6C872 A0A1B6EBX8 A0A1B6C321 A0A1B6DWQ7 A0A1B6D707 A0A1Y1MCV8 A0A0K8TY27 A0A0K8WAU4 F4WEA4 A0A0Q9WUR7 A0A151WEM0 A0A069DWI6 A0A0M4F7E1 A0A0C9RW42 A0A0P4VKQ0 A0A2J7PXR4 A0A146LYW5 A0A0Q9WL53 D0Z7B4 B4MF13 A0A026X480 A0A0P8ZIN0 A0A0J7KUA9 A0A1A9WQC6 E0VLU0 A0A1B6H1P8 A0A0R3P0T0 A0A0L7QYK3 B4H9Q9 A0A1W4WBT9 A0A1W4W145 A0A3B0KUD6 A0A3B0KK07 A0A3B0KQL8
A0A1B6EGA4 A0A1Y1MFW3 A0A1Y1MCG1 A0A2S2Q6M0 A0A2S2QGV5 A0A2H8TNT3 A0A1J1IJL1 T1HJN0 A0A088AF55 A0A067RKX2 J9JQA5 K7IUC4 D6WYU2 E2A256 A0A0C9QNW9 A0A2J7PXS6 A0A0C9R6E0 A0A2J7PXQ6 A0A2J7PXR8 E2C1L4 A0A232FHR6 A0A154PH31 A0A2J7PXS3 A0A2J7PXS1 A0A158N9E7 A0A2J7PXR5 A0A2J7PXU4 A0A2J7PXT3 A0A2J7PXR9 A0A151ILQ7 A0A151IUM6 A0A195FH09 A0A151I1A4 A0A2J7PXT0 A0A2J7PXS8 A0A2J7PXQ5 A0A2J7PXS7 R4WPQ9 A0A2J7PXU1 A0A336L4I9 A0A0A1XRX3 A0A1L8E5U1 A0A1B6CMR6 A0A1B0CD21 A0A1B6DXF3 A0A310S6J9 A0A034VDC1 A0A034VB80 A0A034VCD3 A0A034VDZ5 A0A034VB88 N6U922 A0A1Y1MCG0 A0A1B6CP60 A0A1B6CPR2 A0A1B6C872 A0A1B6EBX8 A0A1B6C321 A0A1B6DWQ7 A0A1B6D707 A0A1Y1MCV8 A0A0K8TY27 A0A0K8WAU4 F4WEA4 A0A0Q9WUR7 A0A151WEM0 A0A069DWI6 A0A0M4F7E1 A0A0C9RW42 A0A0P4VKQ0 A0A2J7PXR4 A0A146LYW5 A0A0Q9WL53 D0Z7B4 B4MF13 A0A026X480 A0A0P8ZIN0 A0A0J7KUA9 A0A1A9WQC6 E0VLU0 A0A1B6H1P8 A0A0R3P0T0 A0A0L7QYK3 B4H9Q9 A0A1W4WBT9 A0A1W4W145 A0A3B0KUD6 A0A3B0KK07 A0A3B0KQL8
PDB
2D8Y
E-value=3.5263e-13,
Score=182
Ontologies
GO
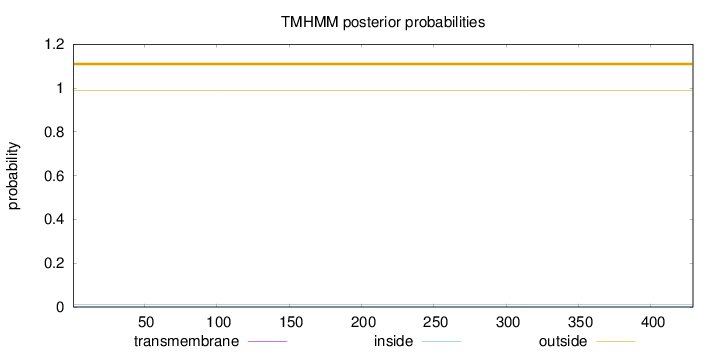
Topology
Length:
429
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01252
outside
1 - 429
Population Genetic Test Statistics
Pi
220.597638
Theta
170.123955
Tajima's D
0.993377
CLR
0.424248
CSRT
0.661166941652917
Interpretation
Uncertain
