Gene
KWMTBOMO16058
Pre Gene Modal
BGIBMGA004584
Annotation
putative_zinc_finger_protein_transcription_factor_lame_duck_[Operophtera_brumata]
Transcription factor
Location in the cell
Nuclear Reliability : 2.758
Sequence
CDS
ATGCGCGTTCACTCTGGACACAAGCCGAACAGATGTCATCACCCGGGCTGCGGGAAGGCCTTCTCCCGTCTGGAGAACCTGAAGATCCACGTGCGGTCCCACACGGGCGAGCGGCCCTACGCCTGCCCGGCGCCGCACTGCAGGAAGGCCTTCTCTAACTCCTCGGATCGAGCGAAACATCAGCGGACCCATTTTAACGCTGTGAGTTTATTGTTATGTTATTTCAAAATTTGTTACGATACTGTTGAATATTAG
Protein
MRVHSGHKPNRCHHPGCGKAFSRLENLKIHVRSHTGERPYACPAPHCRKAFSNSSDRAKHQRTHFNAVSLLLCYFKICYDTVEY
Summary
Uniprot
A0A2A4IZG8
A0A0L7LS16
A0A0N1PGI9
A0A0N1PIA1
H9J4Z4
A0A2H1VZE5
+ More
A0A212ES86 A0A3S2NE44 A0A0C9QUN0 A0A0A9XIN2 A7RU02 A0A1D2MC39 A0A1A7WXR4 E9HVZ7 A0A146WUR1 S7N9F6 D2A690 X1YV67 A0A2P2I8C9 A0A336M7J0 B4JSF1 A0A1X7TZW3 A0A336LZY2 A0A096LX21 F1QLU9 A0A3P8Y6I8 F1QB13 A0A0R4IU95 I3KPR2 A0A3P8NVZ4 A0A3P9DEG4 A0A3B3UN43 A0A3B4F4T0 A1L1Q6 A0A3Q2VLI3 A0A3Q4HB75 A0A3Q0S9C6 B4MB53 A0A0N8DMH8 Q1PHK2 A0A162R3H3 A0A0P5X6X2 A0A0P6JK72 A0A3B4WZL6 A0A0N8E1A9 A0A3P9HM98 A0A1Z5LAI9 U3K7G9 A0A3Q1IV94 A0A3B3CSC6 A0A3P8X0U1 C6KXM5 A0A3P8TYA1 A0A1I8M221 A0A3B4AHS9 A0A3Q4BV79 A0A3Q1CRF7 A0A3Q3LP82 A0A3B5ADY8 B4KBC4 B3LZ98 A0A3Q3GXQ6 A0A0N8CU51 A0A1W4ZLW4 A0A1W4ZLW9 A0A091QP27 A0A093RFM2 A0A1W4V4S4 A0A3B0JLC0 B4R1C1 B3P8H3 A0A091RZC8 B4HF96 A0A094K6C9 B4GYR7 Q962I0 Q7K9G4 A0A091GCN7 D2H2F5 G1LSS7 B4PN27 Q29AW1 A0A0S7I0U6 A0A3B5PWI8 W5LIU3 A0A3B5LZC1 A0A3B3UFB6 A0A2I0U2A1 V4BDH9 G3VFX4 H3AVX8 A0A3B1ILW8 A0A2I0MCD5 A0A3P9PEU5 A0A3B5KEF1 A0A096LST7 A0A091GXG1 C3XY75 A0A091MW11
A0A212ES86 A0A3S2NE44 A0A0C9QUN0 A0A0A9XIN2 A7RU02 A0A1D2MC39 A0A1A7WXR4 E9HVZ7 A0A146WUR1 S7N9F6 D2A690 X1YV67 A0A2P2I8C9 A0A336M7J0 B4JSF1 A0A1X7TZW3 A0A336LZY2 A0A096LX21 F1QLU9 A0A3P8Y6I8 F1QB13 A0A0R4IU95 I3KPR2 A0A3P8NVZ4 A0A3P9DEG4 A0A3B3UN43 A0A3B4F4T0 A1L1Q6 A0A3Q2VLI3 A0A3Q4HB75 A0A3Q0S9C6 B4MB53 A0A0N8DMH8 Q1PHK2 A0A162R3H3 A0A0P5X6X2 A0A0P6JK72 A0A3B4WZL6 A0A0N8E1A9 A0A3P9HM98 A0A1Z5LAI9 U3K7G9 A0A3Q1IV94 A0A3B3CSC6 A0A3P8X0U1 C6KXM5 A0A3P8TYA1 A0A1I8M221 A0A3B4AHS9 A0A3Q4BV79 A0A3Q1CRF7 A0A3Q3LP82 A0A3B5ADY8 B4KBC4 B3LZ98 A0A3Q3GXQ6 A0A0N8CU51 A0A1W4ZLW4 A0A1W4ZLW9 A0A091QP27 A0A093RFM2 A0A1W4V4S4 A0A3B0JLC0 B4R1C1 B3P8H3 A0A091RZC8 B4HF96 A0A094K6C9 B4GYR7 Q962I0 Q7K9G4 A0A091GCN7 D2H2F5 G1LSS7 B4PN27 Q29AW1 A0A0S7I0U6 A0A3B5PWI8 W5LIU3 A0A3B5LZC1 A0A3B3UFB6 A0A2I0U2A1 V4BDH9 G3VFX4 H3AVX8 A0A3B1ILW8 A0A2I0MCD5 A0A3P9PEU5 A0A3B5KEF1 A0A096LST7 A0A091GXG1 C3XY75 A0A091MW11
Pubmed
26227816
26354079
19121390
22118469
25401762
17615350
+ More
27289101 21292972 18362917 19820115 23254933 17994087 23594743 25069045 25186727 16715098 17554307 28528879 29451363 24487278 19609364 25315136 25463417 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 11782407 20010809 17550304 15632085 23542700 25329095 21709235 9215903 23371554 21551351 18563158
27289101 21292972 18362917 19820115 23254933 17994087 23594743 25069045 25186727 16715098 17554307 28528879 29451363 24487278 19609364 25315136 25463417 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 11782407 20010809 17550304 15632085 23542700 25329095 21709235 9215903 23371554 21551351 18563158
EMBL
NWSH01004650
PCG64818.1
JTDY01000218
KOB78237.1
KQ458842
KPJ04901.1
+ More
KQ459890 KPJ19581.1 BABH01025710 ODYU01005403 SOQ46201.1 AGBW02012867 OWR44311.1 RSAL01000078 RVE48730.1 GBYB01007462 GBYB01007463 GBYB01007464 JAG77229.1 JAG77230.1 JAG77231.1 GBHO01024098 JAG19506.1 DS469538 EDO45177.1 LJIJ01001942 ODM90482.1 HADW01009317 SBP10717.1 GL732885 EFX64073.1 GCES01052690 JAR33633.1 KE163605 EPQ12830.1 KQ971346 EFA05809.2 AMQN01000151 IACF01004679 LAB70268.1 UFQT01000504 SSX24839.1 CH916373 EDV94691.1 UFQT01000265 SSX22591.1 AYCK01007466 AYCK01007467 AYCK01007468 CR847832 CT025742 CU468737 AERX01019659 AERX01019660 BC129173 AAI29174.1 CH940656 EDW58324.1 GDIP01019223 JAM84492.1 DQ438880 ABE66437.1 LRGB01000243 KZS20198.1 GDIP01077081 JAM26634.1 GDIQ01016615 JAN78122.1 GDIQ01071596 JAN23141.1 GFJQ02002772 JAW04198.1 AGTO01000642 AB353139 BAH85837.1 CH933806 EDW16852.2 CH902617 EDV43025.1 GDIP01098639 JAM05076.1 KK702719 KFQ28356.1 KL443527 KFW94817.1 OUUW01000007 SPP83025.1 CM000364 EDX14007.1 CH954182 EDV54068.1 KK937871 KFQ48322.1 CH480815 EDW43274.1 KL342558 KFZ53806.1 CH479198 EDW28186.1 AY032609 AY121651 AE014297 AAK39641.1 AAM51978.1 AAN13923.1 AJ311850 CAC51080.1 KL448089 KFO80145.1 GL192442 EFB29153.1 ACTA01058125 ACTA01066125 ACTA01074125 ACTA01082125 ACTA01090125 ACTA01098125 ACTA01106125 CM000160 EDW98082.2 CM000070 EAL27238.3 GBYX01436205 JAO45130.1 KZ506319 PKU40196.1 KB203019 ESO86629.1 AEFK01179653 AEFK01179654 AEFK01179655 AEFK01179656 AEFK01179657 AEFK01179658 AEFK01179659 AEFK01179660 AEFK01179661 AEFK01179662 AFYH01131998 AFYH01131999 AFYH01132000 AFYH01132001 AFYH01132002 AFYH01132003 AFYH01132004 AFYH01132005 AKCR02000021 PKK27341.1 KL513452 KFO87232.1 GG666472 EEN67129.1 KK837583 KFP80774.1
KQ459890 KPJ19581.1 BABH01025710 ODYU01005403 SOQ46201.1 AGBW02012867 OWR44311.1 RSAL01000078 RVE48730.1 GBYB01007462 GBYB01007463 GBYB01007464 JAG77229.1 JAG77230.1 JAG77231.1 GBHO01024098 JAG19506.1 DS469538 EDO45177.1 LJIJ01001942 ODM90482.1 HADW01009317 SBP10717.1 GL732885 EFX64073.1 GCES01052690 JAR33633.1 KE163605 EPQ12830.1 KQ971346 EFA05809.2 AMQN01000151 IACF01004679 LAB70268.1 UFQT01000504 SSX24839.1 CH916373 EDV94691.1 UFQT01000265 SSX22591.1 AYCK01007466 AYCK01007467 AYCK01007468 CR847832 CT025742 CU468737 AERX01019659 AERX01019660 BC129173 AAI29174.1 CH940656 EDW58324.1 GDIP01019223 JAM84492.1 DQ438880 ABE66437.1 LRGB01000243 KZS20198.1 GDIP01077081 JAM26634.1 GDIQ01016615 JAN78122.1 GDIQ01071596 JAN23141.1 GFJQ02002772 JAW04198.1 AGTO01000642 AB353139 BAH85837.1 CH933806 EDW16852.2 CH902617 EDV43025.1 GDIP01098639 JAM05076.1 KK702719 KFQ28356.1 KL443527 KFW94817.1 OUUW01000007 SPP83025.1 CM000364 EDX14007.1 CH954182 EDV54068.1 KK937871 KFQ48322.1 CH480815 EDW43274.1 KL342558 KFZ53806.1 CH479198 EDW28186.1 AY032609 AY121651 AE014297 AAK39641.1 AAM51978.1 AAN13923.1 AJ311850 CAC51080.1 KL448089 KFO80145.1 GL192442 EFB29153.1 ACTA01058125 ACTA01066125 ACTA01074125 ACTA01082125 ACTA01090125 ACTA01098125 ACTA01106125 CM000160 EDW98082.2 CM000070 EAL27238.3 GBYX01436205 JAO45130.1 KZ506319 PKU40196.1 KB203019 ESO86629.1 AEFK01179653 AEFK01179654 AEFK01179655 AEFK01179656 AEFK01179657 AEFK01179658 AEFK01179659 AEFK01179660 AEFK01179661 AEFK01179662 AFYH01131998 AFYH01131999 AFYH01132000 AFYH01132001 AFYH01132002 AFYH01132003 AFYH01132004 AFYH01132005 AKCR02000021 PKK27341.1 KL513452 KFO87232.1 GG666472 EEN67129.1 KK837583 KFP80774.1
Proteomes
UP000218220
UP000037510
UP000053268
UP000053240
UP000005204
UP000007151
+ More
UP000283053 UP000001593 UP000094527 UP000000305 UP000007266 UP000001070 UP000007879 UP000028760 UP000000437 UP000265140 UP000005207 UP000265100 UP000265160 UP000261500 UP000261460 UP000264840 UP000261580 UP000261340 UP000008792 UP000076858 UP000261360 UP000265200 UP000016665 UP000265040 UP000261560 UP000265120 UP000001038 UP000265080 UP000095301 UP000261520 UP000261620 UP000257160 UP000261640 UP000261400 UP000009192 UP000007801 UP000261660 UP000192224 UP000192221 UP000268350 UP000000304 UP000008711 UP000001292 UP000008744 UP000000803 UP000053760 UP000008912 UP000002282 UP000001819 UP000002852 UP000018467 UP000261380 UP000030746 UP000007648 UP000008672 UP000053872 UP000242638 UP000005226 UP000001554
UP000283053 UP000001593 UP000094527 UP000000305 UP000007266 UP000001070 UP000007879 UP000028760 UP000000437 UP000265140 UP000005207 UP000265100 UP000265160 UP000261500 UP000261460 UP000264840 UP000261580 UP000261340 UP000008792 UP000076858 UP000261360 UP000265200 UP000016665 UP000265040 UP000261560 UP000265120 UP000001038 UP000265080 UP000095301 UP000261520 UP000261620 UP000257160 UP000261640 UP000261400 UP000009192 UP000007801 UP000261660 UP000192224 UP000192221 UP000268350 UP000000304 UP000008711 UP000001292 UP000008744 UP000000803 UP000053760 UP000008912 UP000002282 UP000001819 UP000002852 UP000018467 UP000261380 UP000030746 UP000007648 UP000008672 UP000053872 UP000242638 UP000005226 UP000001554
Interpro
Gene 3D
ProteinModelPortal
A0A2A4IZG8
A0A0L7LS16
A0A0N1PGI9
A0A0N1PIA1
H9J4Z4
A0A2H1VZE5
+ More
A0A212ES86 A0A3S2NE44 A0A0C9QUN0 A0A0A9XIN2 A7RU02 A0A1D2MC39 A0A1A7WXR4 E9HVZ7 A0A146WUR1 S7N9F6 D2A690 X1YV67 A0A2P2I8C9 A0A336M7J0 B4JSF1 A0A1X7TZW3 A0A336LZY2 A0A096LX21 F1QLU9 A0A3P8Y6I8 F1QB13 A0A0R4IU95 I3KPR2 A0A3P8NVZ4 A0A3P9DEG4 A0A3B3UN43 A0A3B4F4T0 A1L1Q6 A0A3Q2VLI3 A0A3Q4HB75 A0A3Q0S9C6 B4MB53 A0A0N8DMH8 Q1PHK2 A0A162R3H3 A0A0P5X6X2 A0A0P6JK72 A0A3B4WZL6 A0A0N8E1A9 A0A3P9HM98 A0A1Z5LAI9 U3K7G9 A0A3Q1IV94 A0A3B3CSC6 A0A3P8X0U1 C6KXM5 A0A3P8TYA1 A0A1I8M221 A0A3B4AHS9 A0A3Q4BV79 A0A3Q1CRF7 A0A3Q3LP82 A0A3B5ADY8 B4KBC4 B3LZ98 A0A3Q3GXQ6 A0A0N8CU51 A0A1W4ZLW4 A0A1W4ZLW9 A0A091QP27 A0A093RFM2 A0A1W4V4S4 A0A3B0JLC0 B4R1C1 B3P8H3 A0A091RZC8 B4HF96 A0A094K6C9 B4GYR7 Q962I0 Q7K9G4 A0A091GCN7 D2H2F5 G1LSS7 B4PN27 Q29AW1 A0A0S7I0U6 A0A3B5PWI8 W5LIU3 A0A3B5LZC1 A0A3B3UFB6 A0A2I0U2A1 V4BDH9 G3VFX4 H3AVX8 A0A3B1ILW8 A0A2I0MCD5 A0A3P9PEU5 A0A3B5KEF1 A0A096LST7 A0A091GXG1 C3XY75 A0A091MW11
A0A212ES86 A0A3S2NE44 A0A0C9QUN0 A0A0A9XIN2 A7RU02 A0A1D2MC39 A0A1A7WXR4 E9HVZ7 A0A146WUR1 S7N9F6 D2A690 X1YV67 A0A2P2I8C9 A0A336M7J0 B4JSF1 A0A1X7TZW3 A0A336LZY2 A0A096LX21 F1QLU9 A0A3P8Y6I8 F1QB13 A0A0R4IU95 I3KPR2 A0A3P8NVZ4 A0A3P9DEG4 A0A3B3UN43 A0A3B4F4T0 A1L1Q6 A0A3Q2VLI3 A0A3Q4HB75 A0A3Q0S9C6 B4MB53 A0A0N8DMH8 Q1PHK2 A0A162R3H3 A0A0P5X6X2 A0A0P6JK72 A0A3B4WZL6 A0A0N8E1A9 A0A3P9HM98 A0A1Z5LAI9 U3K7G9 A0A3Q1IV94 A0A3B3CSC6 A0A3P8X0U1 C6KXM5 A0A3P8TYA1 A0A1I8M221 A0A3B4AHS9 A0A3Q4BV79 A0A3Q1CRF7 A0A3Q3LP82 A0A3B5ADY8 B4KBC4 B3LZ98 A0A3Q3GXQ6 A0A0N8CU51 A0A1W4ZLW4 A0A1W4ZLW9 A0A091QP27 A0A093RFM2 A0A1W4V4S4 A0A3B0JLC0 B4R1C1 B3P8H3 A0A091RZC8 B4HF96 A0A094K6C9 B4GYR7 Q962I0 Q7K9G4 A0A091GCN7 D2H2F5 G1LSS7 B4PN27 Q29AW1 A0A0S7I0U6 A0A3B5PWI8 W5LIU3 A0A3B5LZC1 A0A3B3UFB6 A0A2I0U2A1 V4BDH9 G3VFX4 H3AVX8 A0A3B1ILW8 A0A2I0MCD5 A0A3P9PEU5 A0A3B5KEF1 A0A096LST7 A0A091GXG1 C3XY75 A0A091MW11
PDB
2GLI
E-value=4.93999e-15,
Score=191
Ontologies
GO
GO:0005319
GO:0003676
GO:0000977
GO:0006357
GO:0005634
GO:0007417
GO:0000981
GO:0042593
GO:0003323
GO:1990798
GO:0005737
GO:0007525
GO:0007519
GO:0010817
GO:0045944
GO:0007520
GO:0007517
GO:0003700
GO:0006355
GO:0001227
GO:0010454
GO:0001649
GO:0045444
GO:0001228
GO:0046872
GO:0008270
GO:0022857
GO:0001678
GO:0005536
GO:0046835
GO:0006281
GO:0006259
PANTHER
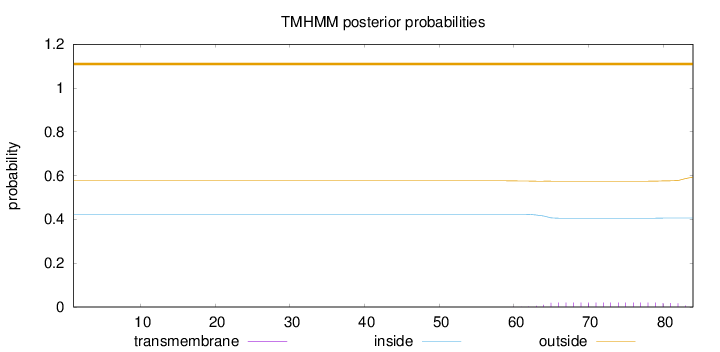
Topology
Length:
84
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.37882
Exp number, first 60 AAs:
0.00041
Total prob of N-in:
0.42384
outside
1 - 84
Population Genetic Test Statistics
Pi
7.805884
Theta
17.309039
Tajima's D
-1.306542
CLR
123.57694
CSRT
0.0820458977051148
Interpretation
Uncertain
