Gene
KWMTBOMO16054 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004586
Annotation
PREDICTED:_trehalase-2_isoform_X1_[Bombyx_mori]
Full name
Trehalase
Alternative Name
Alpha-trehalose glucohydrolase
Alpha,alpha-trehalase
Alpha,alpha-trehalose glucohydrolase
Alpha,alpha-trehalase
Alpha,alpha-trehalose glucohydrolase
Location in the cell
Cytoplasmic Reliability : 1.654
Sequence
CDS
ATGTTTATGGTGTGGCTTTGGGTCGGTTTTGCGATGGCCGCTAGTGTCGTTGCTGATCGTAGCAAACTACCTCCTGCCTGTGATAGTATGATTTACTGCCACGGACCGTTGTTAAACACGGTCCAAATGGCAGGCCTCTACAATGACTCCAAGACGTTTGTGGATATGAAAATTAAAATGTCACCAAAGATCACCTTGGAGCACTTCTATGACATGATGAGCAGGACAGATTCTAATCCGACTAAAGCGGACATACAGGAGTTCGTGAATCAGAACTTCGATCCTGAGGGTTCGGAATTTGAAGACTGGAGGCCCAGTGATTGGAAGCATAACCCTGGATTTCTTGCTAAAATCAAGGATCCGTTATTGCACAAATGGGCATCAGCCTTGAACGATTTATGGCTCGATCTCGGGAGGAAGATGAAGGAAGCAGTCAAAGAGAGTCCAGACTTATACTCCATTATATACGTGGAGCATCCTTTTATTGTTCCAGGTGGTCGTTTCCGTGAGTTCTACTACTGGGACTCGTACTGGATCATCAAAGGTCTGCTGCTCTCGGAGATGAGGACCACAGCGAAGGGCATGGTCAACAACTTACTCAGCATAGTCGATAGATACGGTTTCATACCGAATGGCGGCAGAATTTATTATCTTATGAGATCTCAACCTCCCCTTCTAATACCGATGGTGCAGCTACTAATGGAAGACACCGATGACCTGGAATACCTGAGGGAGCACATCCACACCCTGGACAAAGAGTTCGACTACTGGATGACGAATCACACCATCGAAATTAACCACGAAGGGAAGAAATTGAAAATGGCGATGTACATGGATAATTCTCAGGGTCCGAGGCCGGAGAGCTACAAAGAAGACGTCGACTGTGCTAGACATTTTGACACAACTGAGAAGAAAGAAGAATTGTACGCTGAATTGAAGGCTGCAGCTGAATCCGGCTGGGATTTCTCATCAAGATGGTTTATACTGAACGGAACCAATAAAGGTAATCTAACTAATTTGAAGGTGAGGTCGATAATTCCCGTCGACCTCAACGCGATCTTGTGCTGGAACGCGCAGCTCATGATGGAGTACCACACCCGCCTTCAAAATGAGGAGAAAGCATCTTACTATAGGCGGATACACGATGACTTCATGGAAGCTATCGAAGAGCTATTGTGGCACGAAGACGTGGGAGCTTGGTTGGACTACAGCCTCGAGTCTGGTCGCAGGAGAGACTACTTTTACCCATCCAACCTGTCTCCGCTCTGGGCCGGCAGCTTCGATAAAGCGAGAAAGGATTATTTCGTGAATAGGGTCATCAATTATTTGGACAAAGTTAAGATGGACATCTTCGAAGGCGGAATCCCAACAACGTTCGAACATACCGGCGAACAGTGGGACTATCCCAACGCGTGGCCGCCCCTCCAGTACATCGTCATAATGGGACTAGCCAACACCGGGCACCCGGAGGCGATGAGATACGCCACCGAACTCGCCACTAAGTGGGTGCGGAGTAACTTCGAAGTCTGGAAGCAGAAGGCCGCCATGCTGGAGAAGTACGACGCCACGATTCTGGGTGGGCTGGGTGGCGGCGGAGAATACGTCGTCCAAACCGGTTTCGGATGGAGCAATGGCGTTGTGATGTCACTCCTAAACAGATATGGAGACGTAATCTCGGCAGCCGACAACTTCGGAACGGCGGCGGAATCGGGCGCTGTCTACGGCGCCGGGGTGGGCGCCGGTGGCGTAGTCACGGCGCTCCTAGTGGTCCTAGCTTCTTTGGCTGCTGGAACTTTAGGATTAATGGTGTACAAGAGACGTAAAGATTACGCCGGCTTCTGTAACGGCGAAGTCGACAAACTCATCTCGAAAAAACCCTACACTGAACTCAAGAGTTTGAACGGCGCGTCGAACCAGAAACAGCGGTGA
Protein
MFMVWLWVGFAMAASVVADRSKLPPACDSMIYCHGPLLNTVQMAGLYNDSKTFVDMKIKMSPKITLEHFYDMMSRTDSNPTKADIQEFVNQNFDPEGSEFEDWRPSDWKHNPGFLAKIKDPLLHKWASALNDLWLDLGRKMKEAVKESPDLYSIIYVEHPFIVPGGRFREFYYWDSYWIIKGLLLSEMRTTAKGMVNNLLSIVDRYGFIPNGGRIYYLMRSQPPLLIPMVQLLMEDTDDLEYLREHIHTLDKEFDYWMTNHTIEINHEGKKLKMAMYMDNSQGPRPESYKEDVDCARHFDTTEKKEELYAELKAAAESGWDFSSRWFILNGTNKGNLTNLKVRSIIPVDLNAILCWNAQLMMEYHTRLQNEEKASYYRRIHDDFMEAIEELLWHEDVGAWLDYSLESGRRRDYFYPSNLSPLWAGSFDKARKDYFVNRVINYLDKVKMDIFEGGIPTTFEHTGEQWDYPNAWPPLQYIVIMGLANTGHPEAMRYATELATKWVRSNFEVWKQKAAMLEKYDATILGGLGGGGEYVVQTGFGWSNGVVMSLLNRYGDVISAADNFGTAAESGAVYGAGVGAGGVVTALLVVLASLAAGTLGLMVYKRRKDYAGFCNGEVDKLISKKPYTELKSLNGASNQKQR
Summary
Catalytic Activity
alpha,alpha-trehalose + H2O = alpha-D-glucose + beta-D-glucose
Biophysicochemical Properties
0.66 mM for alpha,alpha-trehalose (in 0.1 M sodium phosphate pH 6.75, at 35 degrees Celsius)
0.52 mM for alpha,alpha-trehalose (in 50 mM sodium phosphate pH 6.6, at 35 degrees Celsius)
0.52 mM for alpha,alpha-trehalose (in 50 mM sodium phosphate pH 6.6, at 35 degrees Celsius)
Subunit
Monomer.
Similarity
Belongs to the glycosyl hydrolase 37 family.
Keywords
Complete proteome
Direct protein sequencing
Glycoprotein
Glycosidase
Hydrolase
Membrane
Reference proteome
Signal
Transmembrane
Transmembrane helix
Feature
chain Trehalase
Uniprot
Q3MV18
H9J4Z6
B5ATV4
A7XZC0
A0A2D0WLR5
A0A0F6PAZ3
+ More
D2KHI9 A0A3G2SGP7 A0A2W1BGC4 A0A1B2AQF4 A3RLQ4 A0A1U9A8C9 A0A212ES68 A0A2R7X5I1 S4UQ18 A0A1B6KL82 A0A1B6KBM1 A0A0A9WUU0 A0A0K8TDD3 A0A146LQW6 K7R0G4 A0A1B6F1K5 A0A288VJ48 I3WEV3 A0A1S6Q350 C0LZJ7 A0A1S3D5V1 A0A1S3D5U6 A0A1S3D5L1 A0A1S3D5M3 I7CKW6 A0A2P8YXZ4 V9I8L8 A0A1D8I2M2 A0A2S2PXE7 A0A1W4WRK7 E1ZVE9 A0A2J7PTL5 C8CGD2 A0A2J7PTK0 A8J4S9 A0A088AUT8 A0A1B6DUA6 A0A1B6EEF7 A0A310SI70 A0A1W4WFC2 A0A2H8TFR3 J9JU09 D6W851 E2BGR0 G9I465 A0A3L8DTI2 V5GU59 A0A1X9IF78 A0A2J7Q3Z7 A0A195BF82 E0VKM4 A0A067R5E2 A0A1B6EC02 A0A0C9S2J2 A0A0C9S020 A0A067QSZ4 A0A026VXI6 A0A195FE62 E9I9Z8 A0A158NBN3 F4X8H6 K7IXR0 A0A0C9REV2 A0A1W4WQA7 A0A336MHM1 A0A2P8YZX5 A0A195DS33 U4UVG1 A0A336MLY4 A0A023F5Y3 T1HKP0 A0A0J7L328 A0A224X931 A0A1Y1LKH8 A0A1Y1LN31 A0A1S6Q338 A0A151WL58 A0A0L7QL65 A0A151IIY2 A0A0A1X8X4 E0W4A8 A0A2A3E7C0 J7EJ73 J9Q3L8 A0A034VU16 A0A1L8E1D6 A0A1B0GQK5 A0A034VSD4 A0A1L8E177 A0A1L8E1T5 A0A034VTZ9 A0A034VRC1 A0A1L8E1B2
D2KHI9 A0A3G2SGP7 A0A2W1BGC4 A0A1B2AQF4 A3RLQ4 A0A1U9A8C9 A0A212ES68 A0A2R7X5I1 S4UQ18 A0A1B6KL82 A0A1B6KBM1 A0A0A9WUU0 A0A0K8TDD3 A0A146LQW6 K7R0G4 A0A1B6F1K5 A0A288VJ48 I3WEV3 A0A1S6Q350 C0LZJ7 A0A1S3D5V1 A0A1S3D5U6 A0A1S3D5L1 A0A1S3D5M3 I7CKW6 A0A2P8YXZ4 V9I8L8 A0A1D8I2M2 A0A2S2PXE7 A0A1W4WRK7 E1ZVE9 A0A2J7PTL5 C8CGD2 A0A2J7PTK0 A8J4S9 A0A088AUT8 A0A1B6DUA6 A0A1B6EEF7 A0A310SI70 A0A1W4WFC2 A0A2H8TFR3 J9JU09 D6W851 E2BGR0 G9I465 A0A3L8DTI2 V5GU59 A0A1X9IF78 A0A2J7Q3Z7 A0A195BF82 E0VKM4 A0A067R5E2 A0A1B6EC02 A0A0C9S2J2 A0A0C9S020 A0A067QSZ4 A0A026VXI6 A0A195FE62 E9I9Z8 A0A158NBN3 F4X8H6 K7IXR0 A0A0C9REV2 A0A1W4WQA7 A0A336MHM1 A0A2P8YZX5 A0A195DS33 U4UVG1 A0A336MLY4 A0A023F5Y3 T1HKP0 A0A0J7L328 A0A224X931 A0A1Y1LKH8 A0A1Y1LN31 A0A1S6Q338 A0A151WL58 A0A0L7QL65 A0A151IIY2 A0A0A1X8X4 E0W4A8 A0A2A3E7C0 J7EJ73 J9Q3L8 A0A034VU16 A0A1L8E1D6 A0A1B0GQK5 A0A034VSD4 A0A1L8E177 A0A1L8E1T5 A0A034VTZ9 A0A034VRC1 A0A1L8E1B2
EC Number
3.2.1.28
Pubmed
EMBL
AB162717
BAE45249.1
BABH01025716
EU872435
ACF94698.1
EU106080
+ More
ABU95354.1 KU977457 ARD05075.1 KJ652558 AJK29980.1 GU211890 ADA63845.1 MG787168 AYO46921.1 KZ150067 PZC74122.1 KU640202 ANY30159.1 EF426723 ABO20845.1 KX010600 ANC68249.1 AGBW02012867 OWR44309.1 KK857312 PTY27036.1 JX312193 AGL34007.2 GEBQ01027776 JAT12201.1 GEBQ01031162 JAT08815.1 GBHO01031362 JAG12242.1 GBRD01002317 JAG63504.1 GDHC01009483 JAQ09146.1 JX024262 AFV79627.1 GECZ01025694 JAS44075.1 KY400004 KY400005 KY400006 AQY15496.1 JQ027051 AFL03410.1 KX371565 AQV08187.1 FJ790320 ACN85421.1 JX204291 AFO54713.1 PYGN01000293 PSN49112.1 JR036191 JR036192 JR036193 JR036194 JR036195 JR036196 AEY56985.1 AEY56986.1 AEY56987.1 AEY56988.1 AEY56989.1 AEY56990.1 KU756286 AOT99589.1 GGMS01000994 MBY70197.1 GL434492 EFN74859.1 NEVH01021237 PNF19664.1 GQ397451 ACV20872.1 PNF19666.1 AB301556 BAF81545.1 GEDC01008038 JAS29260.1 GEDC01000986 JAS36312.1 KQ759866 OAD62478.1 GFXV01001141 MBW12946.1 ABLF02022564 KQ971307 EFA11183.1 GL448204 EFN85130.1 JN565077 AEW67359.1 QOIP01000004 RLU23209.1 GALX01000767 JAB67699.1 KX349225 AOT82130.1 NEVH01018420 PNF23300.1 KQ976500 KYM83241.1 DS235248 EEB13930.1 KK852732 KDR17472.1 GEDC01001832 JAS35466.1 GBYB01015126 JAG84893.1 GBYB01015125 JAG84892.1 KK852980 KDR13013.1 KK107638 EZA48385.1 KQ981673 KYN38309.1 GL761864 EFZ22607.1 ADTU01011229 ADTU01011230 ADTU01011231 GL888932 EGI57245.1 GBYB01015124 JAG84891.1 UFQS01001260 UFQT01001260 SSX10014.1 SSX29736.1 PYGN01000262 PSN49799.1 KQ980581 KYN15329.1 KB632384 ERL94231.1 UFQS01001328 UFQT01001328 SSX10371.1 SSX30059.1 GBBI01002113 JAC16599.1 ACPB03020626 LBMM01000908 KMQ97237.1 GFTR01007580 JAW08846.1 GEZM01054933 GEZM01054930 GEZM01054928 GEZM01054926 JAV73338.1 GEZM01054932 GEZM01054931 GEZM01054929 GEZM01054927 JAV73335.1 KX371563 AQV08185.1 KQ982981 KYQ48550.1 KQ414934 KOC59334.1 KQ977445 KYN02611.1 GBXI01006976 JAD07316.1 DS235886 EEB20464.1 KZ288356 PBC27182.1 HQ231258 ADT91708.1 JQ228448 GAKP01013934 GAKP01013922 GAKP01013920 AFI80736.1 JAC45018.1 GAKP01013914 JAC45038.1 GFDF01001610 JAV12474.1 AJVK01017190 AJVK01017191 GAKP01013928 GAKP01013925 GAKP01013915 JAC45037.1 GFDF01001606 JAV12478.1 GFDF01001609 JAV12475.1 GAKP01013929 JAC45023.1 GAKP01013933 GAKP01013926 JAC45019.1 GFDF01001605 JAV12479.1
ABU95354.1 KU977457 ARD05075.1 KJ652558 AJK29980.1 GU211890 ADA63845.1 MG787168 AYO46921.1 KZ150067 PZC74122.1 KU640202 ANY30159.1 EF426723 ABO20845.1 KX010600 ANC68249.1 AGBW02012867 OWR44309.1 KK857312 PTY27036.1 JX312193 AGL34007.2 GEBQ01027776 JAT12201.1 GEBQ01031162 JAT08815.1 GBHO01031362 JAG12242.1 GBRD01002317 JAG63504.1 GDHC01009483 JAQ09146.1 JX024262 AFV79627.1 GECZ01025694 JAS44075.1 KY400004 KY400005 KY400006 AQY15496.1 JQ027051 AFL03410.1 KX371565 AQV08187.1 FJ790320 ACN85421.1 JX204291 AFO54713.1 PYGN01000293 PSN49112.1 JR036191 JR036192 JR036193 JR036194 JR036195 JR036196 AEY56985.1 AEY56986.1 AEY56987.1 AEY56988.1 AEY56989.1 AEY56990.1 KU756286 AOT99589.1 GGMS01000994 MBY70197.1 GL434492 EFN74859.1 NEVH01021237 PNF19664.1 GQ397451 ACV20872.1 PNF19666.1 AB301556 BAF81545.1 GEDC01008038 JAS29260.1 GEDC01000986 JAS36312.1 KQ759866 OAD62478.1 GFXV01001141 MBW12946.1 ABLF02022564 KQ971307 EFA11183.1 GL448204 EFN85130.1 JN565077 AEW67359.1 QOIP01000004 RLU23209.1 GALX01000767 JAB67699.1 KX349225 AOT82130.1 NEVH01018420 PNF23300.1 KQ976500 KYM83241.1 DS235248 EEB13930.1 KK852732 KDR17472.1 GEDC01001832 JAS35466.1 GBYB01015126 JAG84893.1 GBYB01015125 JAG84892.1 KK852980 KDR13013.1 KK107638 EZA48385.1 KQ981673 KYN38309.1 GL761864 EFZ22607.1 ADTU01011229 ADTU01011230 ADTU01011231 GL888932 EGI57245.1 GBYB01015124 JAG84891.1 UFQS01001260 UFQT01001260 SSX10014.1 SSX29736.1 PYGN01000262 PSN49799.1 KQ980581 KYN15329.1 KB632384 ERL94231.1 UFQS01001328 UFQT01001328 SSX10371.1 SSX30059.1 GBBI01002113 JAC16599.1 ACPB03020626 LBMM01000908 KMQ97237.1 GFTR01007580 JAW08846.1 GEZM01054933 GEZM01054930 GEZM01054928 GEZM01054926 JAV73338.1 GEZM01054932 GEZM01054931 GEZM01054929 GEZM01054927 JAV73335.1 KX371563 AQV08185.1 KQ982981 KYQ48550.1 KQ414934 KOC59334.1 KQ977445 KYN02611.1 GBXI01006976 JAD07316.1 DS235886 EEB20464.1 KZ288356 PBC27182.1 HQ231258 ADT91708.1 JQ228448 GAKP01013934 GAKP01013922 GAKP01013920 AFI80736.1 JAC45018.1 GAKP01013914 JAC45038.1 GFDF01001610 JAV12474.1 AJVK01017190 AJVK01017191 GAKP01013928 GAKP01013925 GAKP01013915 JAC45037.1 GFDF01001606 JAV12478.1 GFDF01001609 JAV12475.1 GAKP01013929 JAC45023.1 GAKP01013933 GAKP01013926 JAC45019.1 GFDF01001605 JAV12479.1
Proteomes
UP000005204
UP000007151
UP000079169
UP000245037
UP000192223
UP000000311
+ More
UP000235965 UP000005203 UP000007819 UP000007266 UP000008237 UP000279307 UP000078540 UP000009046 UP000027135 UP000053097 UP000078541 UP000005205 UP000007755 UP000002358 UP000078492 UP000030742 UP000015103 UP000036403 UP000075809 UP000053825 UP000078542 UP000242457 UP000092462
UP000235965 UP000005203 UP000007819 UP000007266 UP000008237 UP000279307 UP000078540 UP000009046 UP000027135 UP000053097 UP000078541 UP000005205 UP000007755 UP000002358 UP000078492 UP000030742 UP000015103 UP000036403 UP000075809 UP000053825 UP000078542 UP000242457 UP000092462
Pfam
PF01204 Trehalase
Interpro
SUPFAM
SSF48208
SSF48208
Gene 3D
ProteinModelPortal
Q3MV18
H9J4Z6
B5ATV4
A7XZC0
A0A2D0WLR5
A0A0F6PAZ3
+ More
D2KHI9 A0A3G2SGP7 A0A2W1BGC4 A0A1B2AQF4 A3RLQ4 A0A1U9A8C9 A0A212ES68 A0A2R7X5I1 S4UQ18 A0A1B6KL82 A0A1B6KBM1 A0A0A9WUU0 A0A0K8TDD3 A0A146LQW6 K7R0G4 A0A1B6F1K5 A0A288VJ48 I3WEV3 A0A1S6Q350 C0LZJ7 A0A1S3D5V1 A0A1S3D5U6 A0A1S3D5L1 A0A1S3D5M3 I7CKW6 A0A2P8YXZ4 V9I8L8 A0A1D8I2M2 A0A2S2PXE7 A0A1W4WRK7 E1ZVE9 A0A2J7PTL5 C8CGD2 A0A2J7PTK0 A8J4S9 A0A088AUT8 A0A1B6DUA6 A0A1B6EEF7 A0A310SI70 A0A1W4WFC2 A0A2H8TFR3 J9JU09 D6W851 E2BGR0 G9I465 A0A3L8DTI2 V5GU59 A0A1X9IF78 A0A2J7Q3Z7 A0A195BF82 E0VKM4 A0A067R5E2 A0A1B6EC02 A0A0C9S2J2 A0A0C9S020 A0A067QSZ4 A0A026VXI6 A0A195FE62 E9I9Z8 A0A158NBN3 F4X8H6 K7IXR0 A0A0C9REV2 A0A1W4WQA7 A0A336MHM1 A0A2P8YZX5 A0A195DS33 U4UVG1 A0A336MLY4 A0A023F5Y3 T1HKP0 A0A0J7L328 A0A224X931 A0A1Y1LKH8 A0A1Y1LN31 A0A1S6Q338 A0A151WL58 A0A0L7QL65 A0A151IIY2 A0A0A1X8X4 E0W4A8 A0A2A3E7C0 J7EJ73 J9Q3L8 A0A034VU16 A0A1L8E1D6 A0A1B0GQK5 A0A034VSD4 A0A1L8E177 A0A1L8E1T5 A0A034VTZ9 A0A034VRC1 A0A1L8E1B2
D2KHI9 A0A3G2SGP7 A0A2W1BGC4 A0A1B2AQF4 A3RLQ4 A0A1U9A8C9 A0A212ES68 A0A2R7X5I1 S4UQ18 A0A1B6KL82 A0A1B6KBM1 A0A0A9WUU0 A0A0K8TDD3 A0A146LQW6 K7R0G4 A0A1B6F1K5 A0A288VJ48 I3WEV3 A0A1S6Q350 C0LZJ7 A0A1S3D5V1 A0A1S3D5U6 A0A1S3D5L1 A0A1S3D5M3 I7CKW6 A0A2P8YXZ4 V9I8L8 A0A1D8I2M2 A0A2S2PXE7 A0A1W4WRK7 E1ZVE9 A0A2J7PTL5 C8CGD2 A0A2J7PTK0 A8J4S9 A0A088AUT8 A0A1B6DUA6 A0A1B6EEF7 A0A310SI70 A0A1W4WFC2 A0A2H8TFR3 J9JU09 D6W851 E2BGR0 G9I465 A0A3L8DTI2 V5GU59 A0A1X9IF78 A0A2J7Q3Z7 A0A195BF82 E0VKM4 A0A067R5E2 A0A1B6EC02 A0A0C9S2J2 A0A0C9S020 A0A067QSZ4 A0A026VXI6 A0A195FE62 E9I9Z8 A0A158NBN3 F4X8H6 K7IXR0 A0A0C9REV2 A0A1W4WQA7 A0A336MHM1 A0A2P8YZX5 A0A195DS33 U4UVG1 A0A336MLY4 A0A023F5Y3 T1HKP0 A0A0J7L328 A0A224X931 A0A1Y1LKH8 A0A1Y1LN31 A0A1S6Q338 A0A151WL58 A0A0L7QL65 A0A151IIY2 A0A0A1X8X4 E0W4A8 A0A2A3E7C0 J7EJ73 J9Q3L8 A0A034VU16 A0A1L8E1D6 A0A1B0GQK5 A0A034VSD4 A0A1L8E177 A0A1L8E1T5 A0A034VTZ9 A0A034VRC1 A0A1L8E1B2
PDB
5Z6H
E-value=9.22848e-64,
Score=620
Ontologies
GO
PANTHER
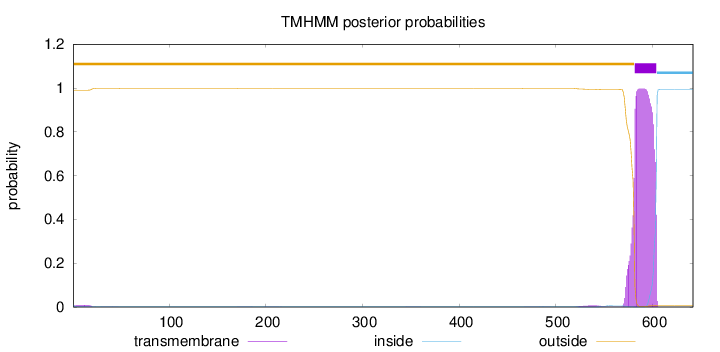
Topology
Subcellular location
Membrane
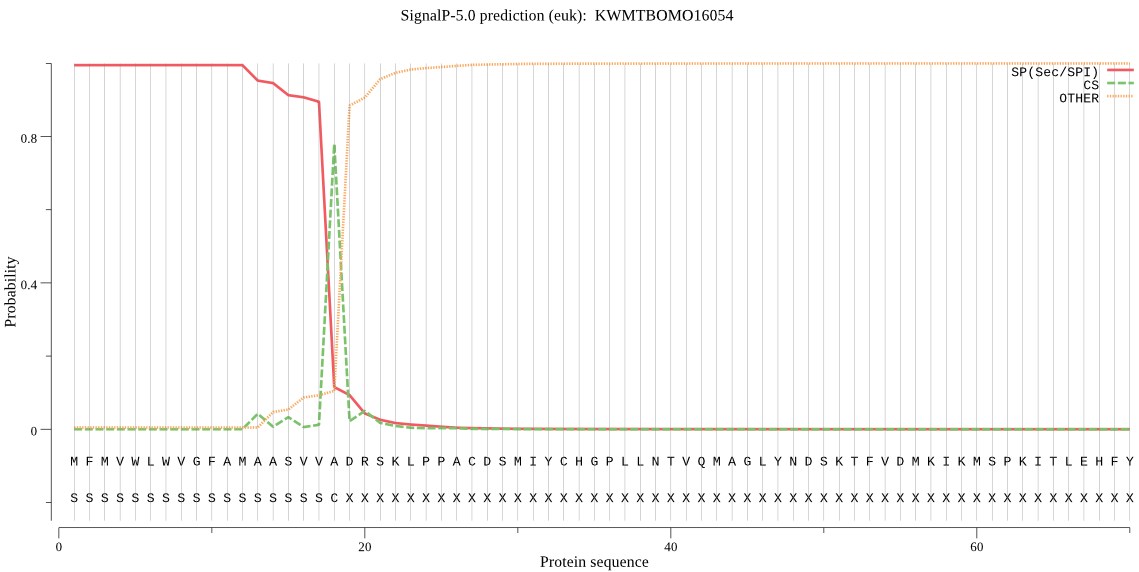
SignalP
Position: 1 - 18,
Likelihood: 0.994838
Length:
642
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
24.23578
Exp number, first 60 AAs:
0.15974
Total prob of N-in:
0.00955
outside
1 - 581
TMhelix
582 - 604
inside
605 - 642
Population Genetic Test Statistics
Pi
316.435468
Theta
160.485393
Tajima's D
3.462168
CLR
0.186435
CSRT
0.992250387480626
Interpretation
Uncertain
