Gene
KWMTBOMO16051
Pre Gene Modal
BGIBMGA004565
Annotation
PREDICTED:_uncharacterized_protein_LOC106720343_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 3.495
Sequence
CDS
ATGACAAAGCCGCGCACACGTCGTCAGTCTCGTCGACAATCCGAGTATTTCGAACCGACAGAGGACGATTCTTCCCCGCCGCCGACTGCATCGGGAAACATTTCCGTCAACGACATGGTCGCCATGATGTCTACGTGGCAACGCAACCAAGAGGAAACGTTTGAGAAACTTCTACATACAGTTCTCAACCAATCGCACCGGTCATCGCCGCCGCCCGCTCAACCTGCACTTGGAAACTTCGCCTCGTGCAAGGCTCGCTACAGCGGAGCGCCAGACGAATCACTTGACGGATTTATAGACGCCGTAGAGGCCTACAAGGAATGTGCAAACGTAAGCGAAACTAATGCACTACGTGGGTTGTCAATGTTGTTAACTCATGAAGCCGCGACATGGTGGCAAGGAGTTAAGACCACCATGAACAAGTGGGACCACGCCCTCACATCCCTAAGAGGTGCGTTTGGTGATAGCCGACCTCCACATCGTATATACAAGGAGCTGTTCGCCAACCCACAAAAAGCAAGTGAGAAAACAGAACTCTTCATCTCTAAAACTCGAGCGCTACTATCACGTCTGCCAAAGGACGATCTGACAACCAAAGTACAGATGGACATGGTATATGGACTACTACATAGTAAAATACGCGAGCGTTTGAGAAGAGAGGAGATTGACACATTTGACACGCTACTCAGAAAGTCTAGAGAGATAGAAGAATCTTGTGACGAGAGTCTATCAACTAGTGGCCTTCAGAGCAAACTGCGCCAACGCCAGTTTCCACCGCCTCGTCACGCCGCAAGCATCGCCGCTGTCCAGGGGTCAACGACCGCGCCACCACTACCACCGCAGAGCGCGCCTAAGCCGCCTGCGCCCGCCGTTGAGTCCAGCGCCCACGGCACCTATCGCCGTTCTCTTCGCTGCTCCGTGGTGCCGCCCCCACCGTCGTCTCCACAACCCCACGGCTCTTCTACTCAGTTCGTGAACGCCAGACGTCGATATTGTTTCTATTGCAAGAACTATGGACATACACGTGAAGAGTGTCGTAGGTTAGCAGCTAAAGGCAAACAGGACATCGATAACGCCGCCGACCCCGTCCCGTCGTGTTACGGATGCAACGAGAAAGGTGTCACTCGATCACAGTGTGCTAAATGCAATTCGGACTTCTCTGCGTGTGAATACGACGGTCCGAAAGGCGCGCTGATGGGAATTTACGGTACAAGGTACTCCACCACTGTTATCAAATCATCTTGTAATAAGGATGAGACCCAGATGTCCGACTCGTACATGTGGAAAGAACAGATTCACACTCTAATGAAGGGCGGAGAGAGCGAGTCCGACATATCGGACTCAGAGCATTTCCTCACAAAGGGCGCCTTCCTTCTCACTCCCTCCCACGGCCTCAGCATGGAAAAAGCCTCCGCCATCTGTGAGAAGATGGAGTTCAAGGGATGCTTCTCCCTGACGAAGACTGCTACTGGAATACTCTTTAAGTTTTCCAGCGTCGATGACTATCAGATGGTATTCAAGAAAGGTTTCCATAAAGTTACCGGATCGCGGTTCTATAGGAAGATCGCTATTCCGTGCAGACCGCAGAAGACCTTTACCATTTACGTATACGACATTCCCGATGAGATCCCAGAGGAGGATGTGCGCCACGCTTTGTACAAATTTACTTCGGTCGTTGAGGTCGTCAGGCTGTATTACTCTGGATCCCCAAAAGACGGGAACACTGGTACATCGACTCCAAAACCAGAGAAGACCTCGTCCCGAGCTCTCACGACTATTGATGGAGAGCTCGTGAGCAGCATGGTGCCGAAGGAAGCCACGAGCGTGGTCCGTGTCACTCTCGCCAACATCGAGGAAGCCAACATTCTGCTGCAAAACGGCCTGGATTTCTACGGAGCTACGTTCTTTCCAACCGAATTAGCTACCCCAACCCAGGCAGCTAAGTTGATTAAGCCTAGCAACAGAGTAACTGGAGGTACCGTGTCAGCCAGAGTCCGTGATTTGCTCCCGGTTTTCGACCAGCAAGGATTCGCCAAGTTCGCTCCACCGGCCTCTAGACTGGTGAAGCCACGTTCCAAGTGA
Protein
MTKPRTRRQSRRQSEYFEPTEDDSSPPPTASGNISVNDMVAMMSTWQRNQEETFEKLLHTVLNQSHRSSPPPAQPALGNFASCKARYSGAPDESLDGFIDAVEAYKECANVSETNALRGLSMLLTHEAATWWQGVKTTMNKWDHALTSLRGAFGDSRPPHRIYKELFANPQKASEKTELFISKTRALLSRLPKDDLTTKVQMDMVYGLLHSKIRERLRREEIDTFDTLLRKSREIEESCDESLSTSGLQSKLRQRQFPPPRHAASIAAVQGSTTAPPLPPQSAPKPPAPAVESSAHGTYRRSLRCSVVPPPPSSPQPHGSSTQFVNARRRYCFYCKNYGHTREECRRLAAKGKQDIDNAADPVPSCYGCNEKGVTRSQCAKCNSDFSACEYDGPKGALMGIYGTRYSTTVIKSSCNKDETQMSDSYMWKEQIHTLMKGGESESDISDSEHFLTKGAFLLTPSHGLSMEKASAICEKMEFKGCFSLTKTATGILFKFSSVDDYQMVFKKGFHKVTGSRFYRKIAIPCRPQKTFTIYVYDIPDEIPEEDVRHALYKFTSVVEVVRLYYSGSPKDGNTGTSTPKPEKTSSRALTTIDGELVSSMVPKEATSVVRVTLANIEEANILLQNGLDFYGATFFPTELATPTQAAKLIKPSNRVTGGTVSARVRDLLPVFDQQGFAKFAPPASRLVKPRSK
Summary
Uniprot
ProteinModelPortal
Ontologies
GO
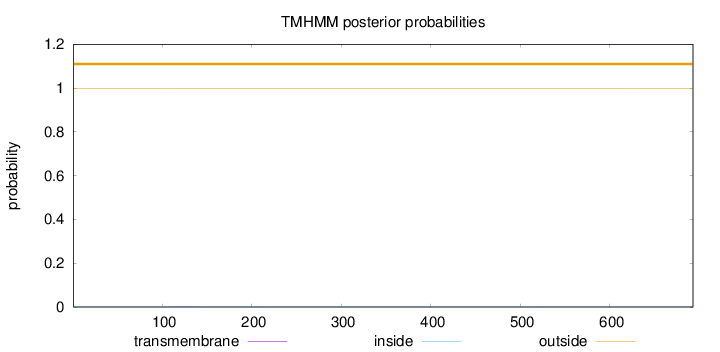
Topology
Length:
693
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00991999999999998
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.00345
outside
1 - 693
Population Genetic Test Statistics
Pi
350.892092
Theta
178.233382
Tajima's D
3.257576
CLR
105.902143
CSRT
0.988800559972001
Interpretation
Uncertain
