Gene
KWMTBOMO16049
Pre Gene Modal
BGIBMGA004563
Annotation
PREDICTED:_facilitated_trehalose_transporter_Tret1-2_homolog_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.986
Sequence
CDS
ATGTATATTTCAGCTTCAATCGCAACCCTACCGGTGGCTCCTGCCTGTCTTATATCTGGATGGATATTTGAAAGATTTGGAAGAAGACCAGCTCTATTCATCACATGTGCCCCGTTTCTACTTGGATGGGCTCTCATAATAAAAGCAAACAACCTCACCCTCATGCTGTGCGGTAGATTTTTCACAGGGATGTGCACAGGATTGATAGGTCCTTTGGCATCGGTCTACATAGGAGAAACTACGGAACCGAAGTATAGAGGTCTCTTCCTCGCTGCCATTTCATTAGCAATGGCATCAGGAATGTTTTTCTCGCATATTATCGGCACATTTATATCATGGCAGTGGACTGCAGTCATCTGCGCTCTGTTGCCAATATTAAGTCTAGTTTTACTAGTTTTTGTACCAGAATCTCCCACTTGGTTAATATCAAGAGGTCACATTGAGGAGGGTGCCAAGGTTTTTTACTGGTTAAGAGGGTATAGTGATGAAGCCAAAGATGAATTGAAAGATATAATCGATCGTAAATTAGCAAACGATGATGCCCCAGTAGAAACTTGGAAAGACAAAATTCGTTATTATAAATGCCCGGAATTTATAAAGCCGTTAGTGATTATGATATTCTTTTTCTCAATGTCCCAATTTGCCGGCGCAAATGCAGTTGCTTTCTATTCTGTTGAAATTATCAAAAAGGCTTTAGGAAAAGATATTGATCAATATATGGCGATGCTCCTAATAGATTTTCTGAGATTGTTAGCGTCAGTAGCAGCCTGTATCATATGCAAACAATACGGGCGAAGACCTGTATGTTTTGTTAGCGGAGGCTTGACAACAATTTCAATGGCAGGACTATCAATATTTTTGTATCTGAGACCAGAAAATATGGCTTGGATACCCCTATCATGCTTCATGCTCTACATTTGTGCGATTTCAATTGGTCTAGTTCCGTTGCCCTGGATGATGTGTGGGGAGATATTTCCGACTAAGGTTCGAGGCTTAGGTTCGGGAGTTAGTTCAGCTGTGGGTTTGGTCTCTGTATTTGTAGTAGTTAAAACCGCACCAGGCATGATGTCTCACTTGGGACAAGTGTTTACTTTCTCGTTTTACGGAGCGATCGCCTTTATTGGAAGCATCATATTATTCTTTGTTCTCCCCGAGACCAAGGGAAGGAGTCTACAGGAGATTGAAGACAAATTTAAAAATCAAAAGAGTAATGGCTCAAAGTTTTGA
Protein
MYISASIATLPVAPACLISGWIFERFGRRPALFITCAPFLLGWALIIKANNLTLMLCGRFFTGMCTGLIGPLASVYIGETTEPKYRGLFLAAISLAMASGMFFSHIIGTFISWQWTAVICALLPILSLVLLVFVPESPTWLISRGHIEEGAKVFYWLRGYSDEAKDELKDIIDRKLANDDAPVETWKDKIRYYKCPEFIKPLVIMIFFFSMSQFAGANAVAFYSVEIIKKALGKDIDQYMAMLLIDFLRLLASVAACIICKQYGRRPVCFVSGGLTTISMAGLSIFLYLRPENMAWIPLSCFMLYICAISIGLVPLPWMMCGEIFPTKVRGLGSGVSSAVGLVSVFVVVKTAPGMMSHLGQVFTFSFYGAIAFIGSIILFFVLPETKGRSLQEIEDKFKNQKSNGSKF
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9J4X3
H9J4X2
E0X906
A0A0N1IPZ8
A0A194QJ63
A0A212FCB5
+ More
H9J4X4 A0A2A4JU31 A0A2H1WFM1 A0A3S2NLE7 A0A2W1BDG5 A0A0L7KM51 A0A1L8DF47 A0A1L8DFA5 A0A1B0CWH0 A0A1B0DPM5 A0A139WGJ0 Q17LS5 A0A182WKH8 A0A182U3G7 A0A182MLS9 A0A182GCQ0 A0A182HWW7 A0A182PHI4 A0A182GTB3 A0A182XN09 A0A1L8DDP6 A0A1L8DDS7 Q7Q6Y6 A0A1S4GQI2 A0A182LJ94 A0A182VI49 A0A182KD28 A0A2P8YLG4 B0W5I2 A0A182NHP5 A0A182IUY7 A0A182Y162 A0A182RRX2 U4UB37 B0W5I1 A0A084VC86 A0A182QST8 A0A194QEH9 A0A1S3CXC8 A0A0A8J8L7 A0A182FDK9 A0A194R9K8 W5JEJ5 A0A2J7RF76 A0A2J7RF66 A0A336MJD0 A0A336M7L2 H9IV67 N6UG74 A0A067QTU4 A0A212ENJ2 A0A1S3CXE4 A0A0L7L7N1 A0A1J1IVY8 A0A2A4IVM9 D4AHX7 A0A336MDU0 A0A1B6CYD7 V9I993 A0A0L7LRT2 V9IBH4 A0A212F9B1 H9J4X7 A0A088AM86 A0A0L7RCV8 V9I974 A0A0N1IPS2 A0A2A3EIY5 A0A2H1V7H3 A0A194QHU0 A0A067RPD0 E2C3F3 A0A2P8XGE2 A0A154P1X7 A0A310SDF5 A0A2A4JTG8 A0A2J7RDW9 A0A2J7RDW1 A0A1B6GJS3 E2ALH5 A0A023PNK9 A0A1B6MC78 A0A151WZ42 K7J6E5 A0A1B6J3Z0 A0A0N1I9K7 A0A232ESL4 T1I127 F4X525 A0A1L8DF63 A0A0N7Z8N1 A0A195EZV9 A0A212F9C9 A0A2H1WF43
H9J4X4 A0A2A4JU31 A0A2H1WFM1 A0A3S2NLE7 A0A2W1BDG5 A0A0L7KM51 A0A1L8DF47 A0A1L8DFA5 A0A1B0CWH0 A0A1B0DPM5 A0A139WGJ0 Q17LS5 A0A182WKH8 A0A182U3G7 A0A182MLS9 A0A182GCQ0 A0A182HWW7 A0A182PHI4 A0A182GTB3 A0A182XN09 A0A1L8DDP6 A0A1L8DDS7 Q7Q6Y6 A0A1S4GQI2 A0A182LJ94 A0A182VI49 A0A182KD28 A0A2P8YLG4 B0W5I2 A0A182NHP5 A0A182IUY7 A0A182Y162 A0A182RRX2 U4UB37 B0W5I1 A0A084VC86 A0A182QST8 A0A194QEH9 A0A1S3CXC8 A0A0A8J8L7 A0A182FDK9 A0A194R9K8 W5JEJ5 A0A2J7RF76 A0A2J7RF66 A0A336MJD0 A0A336M7L2 H9IV67 N6UG74 A0A067QTU4 A0A212ENJ2 A0A1S3CXE4 A0A0L7L7N1 A0A1J1IVY8 A0A2A4IVM9 D4AHX7 A0A336MDU0 A0A1B6CYD7 V9I993 A0A0L7LRT2 V9IBH4 A0A212F9B1 H9J4X7 A0A088AM86 A0A0L7RCV8 V9I974 A0A0N1IPS2 A0A2A3EIY5 A0A2H1V7H3 A0A194QHU0 A0A067RPD0 E2C3F3 A0A2P8XGE2 A0A154P1X7 A0A310SDF5 A0A2A4JTG8 A0A2J7RDW9 A0A2J7RDW1 A0A1B6GJS3 E2ALH5 A0A023PNK9 A0A1B6MC78 A0A151WZ42 K7J6E5 A0A1B6J3Z0 A0A0N1I9K7 A0A232ESL4 T1I127 F4X525 A0A1L8DF63 A0A0N7Z8N1 A0A195EZV9 A0A212F9C9 A0A2H1WF43
Pubmed
EMBL
BABH01025723
BABH01025724
GQ871756
ADM86147.1
KQ460077
KPJ17883.1
+ More
KQ458671 KPJ05557.1 AGBW02009205 OWR51386.1 NWSH01000578 PCG75535.1 ODYU01008354 SOQ51878.1 RSAL01000045 RVE50591.1 KZ150249 PZC71834.1 JTDY01008796 KOB64392.1 GFDF01009026 JAV05058.1 GFDF01009027 JAV05057.1 AJWK01032461 AJVK01008157 KQ971345 KYB27009.1 CH477213 EAT47626.1 AXCM01002831 JXUM01009472 KQ560284 KXJ83288.1 APCN01001553 JXUM01086634 KQ563580 KXJ73638.1 GFDF01009485 JAV04599.1 GFDF01009486 JAV04598.1 AAAB01008960 EAA11457.4 PYGN01000511 PSN45090.1 DS231843 EDS35465.1 KB632225 ERL90272.1 EDS35464.1 ATLV01010625 KE524589 KFB35580.1 AXCN02000368 KQ459299 KPJ01871.1 AB901078 BAQ02378.1 KQ460761 KPJ12541.1 ADMH02001735 ETN61249.1 NEVH01004413 PNF39486.1 PNF39483.1 UFQT01001316 SSX30005.1 UFQT01000224 SSX22028.1 BABH01000587 APGK01036695 APGK01036696 KB740941 ENN77647.1 KK853474 KDR07368.1 AGBW02013662 OWR43027.1 JTDY01002470 KOB71349.1 CVRI01000059 CRL03302.1 NWSH01006268 PCG63546.1 AB550005 BAI83426.1 UFQT01001016 SSX28504.1 GEDC01018890 JAS18408.1 JR037229 AEY57650.1 JTDY01000230 KOB78170.1 JR037228 AEY57649.1 AGBW02009614 OWR50324.1 BABH01025717 KQ414615 KOC68688.1 JR037227 AEY57648.1 KPJ17881.1 KZ288248 PBC31149.1 ODYU01001065 SOQ36788.1 KQ458981 KPJ04505.1 KK852548 KDR21599.1 GL452320 EFN77491.1 PYGN01002224 PSN31080.1 KQ434796 KZC05843.1 KQ762882 OAD55364.1 NWSH01000704 PCG74682.1 NEVH01005280 PNF39024.1 PNF39025.1 GECZ01007067 JAS62702.1 GL440607 EFN65733.1 KJ439227 KZ150491 AHX25878.1 PZC70688.1 GEBQ01006488 JAT33489.1 KQ982649 KYQ52971.1 GECU01013815 JAS93891.1 KPJ17880.1 NNAY01002414 OXU21332.1 ACPB03016561 GL888680 EGI58482.1 GFDF01009003 JAV05081.1 GDKW01002883 JAI53712.1 KQ981905 KYN33447.1 OWR50323.1 ODYU01008270 SOQ51705.1
KQ458671 KPJ05557.1 AGBW02009205 OWR51386.1 NWSH01000578 PCG75535.1 ODYU01008354 SOQ51878.1 RSAL01000045 RVE50591.1 KZ150249 PZC71834.1 JTDY01008796 KOB64392.1 GFDF01009026 JAV05058.1 GFDF01009027 JAV05057.1 AJWK01032461 AJVK01008157 KQ971345 KYB27009.1 CH477213 EAT47626.1 AXCM01002831 JXUM01009472 KQ560284 KXJ83288.1 APCN01001553 JXUM01086634 KQ563580 KXJ73638.1 GFDF01009485 JAV04599.1 GFDF01009486 JAV04598.1 AAAB01008960 EAA11457.4 PYGN01000511 PSN45090.1 DS231843 EDS35465.1 KB632225 ERL90272.1 EDS35464.1 ATLV01010625 KE524589 KFB35580.1 AXCN02000368 KQ459299 KPJ01871.1 AB901078 BAQ02378.1 KQ460761 KPJ12541.1 ADMH02001735 ETN61249.1 NEVH01004413 PNF39486.1 PNF39483.1 UFQT01001316 SSX30005.1 UFQT01000224 SSX22028.1 BABH01000587 APGK01036695 APGK01036696 KB740941 ENN77647.1 KK853474 KDR07368.1 AGBW02013662 OWR43027.1 JTDY01002470 KOB71349.1 CVRI01000059 CRL03302.1 NWSH01006268 PCG63546.1 AB550005 BAI83426.1 UFQT01001016 SSX28504.1 GEDC01018890 JAS18408.1 JR037229 AEY57650.1 JTDY01000230 KOB78170.1 JR037228 AEY57649.1 AGBW02009614 OWR50324.1 BABH01025717 KQ414615 KOC68688.1 JR037227 AEY57648.1 KPJ17881.1 KZ288248 PBC31149.1 ODYU01001065 SOQ36788.1 KQ458981 KPJ04505.1 KK852548 KDR21599.1 GL452320 EFN77491.1 PYGN01002224 PSN31080.1 KQ434796 KZC05843.1 KQ762882 OAD55364.1 NWSH01000704 PCG74682.1 NEVH01005280 PNF39024.1 PNF39025.1 GECZ01007067 JAS62702.1 GL440607 EFN65733.1 KJ439227 KZ150491 AHX25878.1 PZC70688.1 GEBQ01006488 JAT33489.1 KQ982649 KYQ52971.1 GECU01013815 JAS93891.1 KPJ17880.1 NNAY01002414 OXU21332.1 ACPB03016561 GL888680 EGI58482.1 GFDF01009003 JAV05081.1 GDKW01002883 JAI53712.1 KQ981905 KYN33447.1 OWR50323.1 ODYU01008270 SOQ51705.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000218220
UP000283053
+ More
UP000037510 UP000092461 UP000092462 UP000007266 UP000008820 UP000075920 UP000075902 UP000075883 UP000069940 UP000249989 UP000075840 UP000075885 UP000076407 UP000007062 UP000075882 UP000075903 UP000075881 UP000245037 UP000002320 UP000075884 UP000075880 UP000076408 UP000075900 UP000030742 UP000030765 UP000075886 UP000079169 UP000069272 UP000000673 UP000235965 UP000019118 UP000027135 UP000183832 UP000005203 UP000053825 UP000242457 UP000008237 UP000076502 UP000000311 UP000075809 UP000002358 UP000215335 UP000015103 UP000007755 UP000078541
UP000037510 UP000092461 UP000092462 UP000007266 UP000008820 UP000075920 UP000075902 UP000075883 UP000069940 UP000249989 UP000075840 UP000075885 UP000076407 UP000007062 UP000075882 UP000075903 UP000075881 UP000245037 UP000002320 UP000075884 UP000075880 UP000076408 UP000075900 UP000030742 UP000030765 UP000075886 UP000079169 UP000069272 UP000000673 UP000235965 UP000019118 UP000027135 UP000183832 UP000005203 UP000053825 UP000242457 UP000008237 UP000076502 UP000000311 UP000075809 UP000002358 UP000215335 UP000015103 UP000007755 UP000078541
Pfam
PF00083 Sugar_tr
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9J4X3
H9J4X2
E0X906
A0A0N1IPZ8
A0A194QJ63
A0A212FCB5
+ More
H9J4X4 A0A2A4JU31 A0A2H1WFM1 A0A3S2NLE7 A0A2W1BDG5 A0A0L7KM51 A0A1L8DF47 A0A1L8DFA5 A0A1B0CWH0 A0A1B0DPM5 A0A139WGJ0 Q17LS5 A0A182WKH8 A0A182U3G7 A0A182MLS9 A0A182GCQ0 A0A182HWW7 A0A182PHI4 A0A182GTB3 A0A182XN09 A0A1L8DDP6 A0A1L8DDS7 Q7Q6Y6 A0A1S4GQI2 A0A182LJ94 A0A182VI49 A0A182KD28 A0A2P8YLG4 B0W5I2 A0A182NHP5 A0A182IUY7 A0A182Y162 A0A182RRX2 U4UB37 B0W5I1 A0A084VC86 A0A182QST8 A0A194QEH9 A0A1S3CXC8 A0A0A8J8L7 A0A182FDK9 A0A194R9K8 W5JEJ5 A0A2J7RF76 A0A2J7RF66 A0A336MJD0 A0A336M7L2 H9IV67 N6UG74 A0A067QTU4 A0A212ENJ2 A0A1S3CXE4 A0A0L7L7N1 A0A1J1IVY8 A0A2A4IVM9 D4AHX7 A0A336MDU0 A0A1B6CYD7 V9I993 A0A0L7LRT2 V9IBH4 A0A212F9B1 H9J4X7 A0A088AM86 A0A0L7RCV8 V9I974 A0A0N1IPS2 A0A2A3EIY5 A0A2H1V7H3 A0A194QHU0 A0A067RPD0 E2C3F3 A0A2P8XGE2 A0A154P1X7 A0A310SDF5 A0A2A4JTG8 A0A2J7RDW9 A0A2J7RDW1 A0A1B6GJS3 E2ALH5 A0A023PNK9 A0A1B6MC78 A0A151WZ42 K7J6E5 A0A1B6J3Z0 A0A0N1I9K7 A0A232ESL4 T1I127 F4X525 A0A1L8DF63 A0A0N7Z8N1 A0A195EZV9 A0A212F9C9 A0A2H1WF43
H9J4X4 A0A2A4JU31 A0A2H1WFM1 A0A3S2NLE7 A0A2W1BDG5 A0A0L7KM51 A0A1L8DF47 A0A1L8DFA5 A0A1B0CWH0 A0A1B0DPM5 A0A139WGJ0 Q17LS5 A0A182WKH8 A0A182U3G7 A0A182MLS9 A0A182GCQ0 A0A182HWW7 A0A182PHI4 A0A182GTB3 A0A182XN09 A0A1L8DDP6 A0A1L8DDS7 Q7Q6Y6 A0A1S4GQI2 A0A182LJ94 A0A182VI49 A0A182KD28 A0A2P8YLG4 B0W5I2 A0A182NHP5 A0A182IUY7 A0A182Y162 A0A182RRX2 U4UB37 B0W5I1 A0A084VC86 A0A182QST8 A0A194QEH9 A0A1S3CXC8 A0A0A8J8L7 A0A182FDK9 A0A194R9K8 W5JEJ5 A0A2J7RF76 A0A2J7RF66 A0A336MJD0 A0A336M7L2 H9IV67 N6UG74 A0A067QTU4 A0A212ENJ2 A0A1S3CXE4 A0A0L7L7N1 A0A1J1IVY8 A0A2A4IVM9 D4AHX7 A0A336MDU0 A0A1B6CYD7 V9I993 A0A0L7LRT2 V9IBH4 A0A212F9B1 H9J4X7 A0A088AM86 A0A0L7RCV8 V9I974 A0A0N1IPS2 A0A2A3EIY5 A0A2H1V7H3 A0A194QHU0 A0A067RPD0 E2C3F3 A0A2P8XGE2 A0A154P1X7 A0A310SDF5 A0A2A4JTG8 A0A2J7RDW9 A0A2J7RDW1 A0A1B6GJS3 E2ALH5 A0A023PNK9 A0A1B6MC78 A0A151WZ42 K7J6E5 A0A1B6J3Z0 A0A0N1I9K7 A0A232ESL4 T1I127 F4X525 A0A1L8DF63 A0A0N7Z8N1 A0A195EZV9 A0A212F9C9 A0A2H1WF43
PDB
4LDS
E-value=9.79403e-24,
Score=273
Ontologies
GO
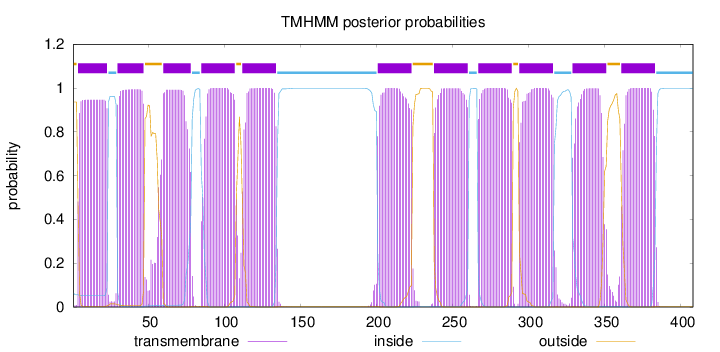
Topology
Length:
408
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
236.3346
Exp number, first 60 AAs:
40.61484
Total prob of N-in:
0.06413
POSSIBLE N-term signal
sequence
outside
1 - 3
TMhelix
4 - 23
inside
24 - 29
TMhelix
30 - 47
outside
48 - 59
TMhelix
60 - 78
inside
79 - 84
TMhelix
85 - 107
outside
108 - 111
TMhelix
112 - 134
inside
135 - 200
TMhelix
201 - 223
outside
224 - 237
TMhelix
238 - 260
inside
261 - 266
TMhelix
267 - 289
outside
290 - 293
TMhelix
294 - 316
inside
317 - 328
TMhelix
329 - 351
outside
352 - 360
TMhelix
361 - 383
inside
384 - 408
Population Genetic Test Statistics
Pi
379.492678
Theta
212.127066
Tajima's D
2.484028
CLR
3.718326
CSRT
0.942502874856257
Interpretation
Uncertain
