Pre Gene Modal
BGIBMGA004550
Annotation
PREDICTED:_epidermal_growth_factor_receptor_kinase_substrate_8-like_isoform_X3_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.525
Sequence
CDS
ATGTCCGCGCAATTGGGCCGCGACAGAGAAAGAGAAAGGGATAGAGACAGAGACCGTAGCGGCTCTGGGGGCGAAAGAGACGACGCTTCATCCACAGGCTCCGAGCGTCTATACGAACAAGACATCGCCATATTGAACCGTTGCTTCGACGACATAGAGAAGTTCATAGCAAGATTACAGCATGCGGCTGCAGCTTCAAGGGAGTTGGAGAGGAGGAGAAGATCGCGAGGAGGCAAGCGAGCGGCGGCTGGTGAAGGAATGCTCGCAATGAGAACCCGTCCTCCGCCGGAGCGCGATTTTGTCGATGTCCTGCAGAAATTTAAACTGTCCTTCAATCTGTTGGCACGTTTGAGAGCTCACATCCATGACCCCAATGCTCCGGAGTTGGTTCATTTCTTGTTCACACCGCTGGCGCTTATTGTGGACGCAGCACAGGACGTCGCCGACGGCCGTCTTCCAGCTAGGGTTGTGCAACCTCTCCTCACGAGGGAGGCCTTGAACCTTCTGGCGAATTGCGTGACAAGCAAAGAAACTGAACTGTGGCACTCATTAGGAGACGCTTGGCTGATACCCAGGGAGCAATGGAAAACACAGATACCACCATATCAACCGGTTTTCATGGACGGGTGGACTCCAGATTACCAAGTCGATGACCAACCTTTACGCAGGTCATCGCCCAGACGCAGCGAGCTGGGCAGGGGTACAATGGAACGGGGCGTGGAAAGGAGCGAGGGCGGGCAGGTGGCGGGTTTCACGTTCGAACGAGAGGAGCCCGAGATATACAACGAGCAATACGCTCCGTTCGCCAGAAATAACTTGAGACCACCAATCGGCCGCGAAGATTCGGGCTCCGCTGCCTCATCGCCCGACAGAGAGCCCCCGTATCGCCCCGCAGACCGAGATGAAGATCTGGGTGAAGCGTGGGCCCGGGGCGTTGCGGCCCGGGGAGGTCGCGTTGTGCGCGTCACCTACCCTCGCACTGCGAATAACGATAAAGAGTTAACTGTCGTGAGGGGAGAGTATTTAGAGGTTCTAGACGATTCGAGAAAGTGGTGGAAGGCTCGTAACAGACGCGGCGTGTCAGCGCACGTGCCACACACCATCGTGGCTCCCGCGTTTTCGCCGCCTTCGTCACCACACCTATACCCTAATCCGATATACACACATTACCAGGATGGCGGGGGGAGAGCTTCGGGCGGCAGCAGTCCCACAGCGGCACCGGAGAAACCAGCGCCAAAGATTGAGGCGGCTGCTCCACCGCCCCCGCCGCCGCCTCCTCCACCCCCGGAGCCGCCGTCACCGCCACCCATCAAATCTGATACTTTGAAATCCACCAAATCGATAATGAGTACTGGCGGCAGTCTCCACGATGAACTGAAGCTAGTACTGCCCCAGATACAGCAAAGAAGAAACAAACTCGACATCAAGAAGACTCCAGATATATTCATTGATCAGAAATCGAATCCGGACGAGGTTGTGGAATGGCTCGAAGCAAAGGGTTTCAGCAATGCCGCCCAACGCCAGCTGAGGATGTCGGGACACCAGCTATTCGCGCTGTCCAGATCTCAGCTGGAGAGGGCACTGGGCCAGGATGAGGGGAAACGACTGTACAGCCAAATTTTGGTCCAGAGGAACGTGTCTGGGTACAAGACGACCTCGGCATCAGAGCTTCAGAGCATCCTGAGGAAAGTACGAGAGAAAGTCGAAGTCTCATGA
Protein
MSAQLGRDRERERDRDRDRSGSGGERDDASSTGSERLYEQDIAILNRCFDDIEKFIARLQHAAAASRELERRRRSRGGKRAAAGEGMLAMRTRPPPERDFVDVLQKFKLSFNLLARLRAHIHDPNAPELVHFLFTPLALIVDAAQDVADGRLPARVVQPLLTREALNLLANCVTSKETELWHSLGDAWLIPREQWKTQIPPYQPVFMDGWTPDYQVDDQPLRRSSPRRSELGRGTMERGVERSEGGQVAGFTFEREEPEIYNEQYAPFARNNLRPPIGREDSGSAASSPDREPPYRPADRDEDLGEAWARGVAARGGRVVRVTYPRTANNDKELTVVRGEYLEVLDDSRKWWKARNRRGVSAHVPHTIVAPAFSPPSSPHLYPNPIYTHYQDGGGRASGGSSPTAAPEKPAPKIEAAAPPPPPPPPPPPEPPSPPPIKSDTLKSTKSIMSTGGSLHDELKLVLPQIQQRRNKLDIKKTPDIFIDQKSNPDEVVEWLEAKGFSNAAQRQLRMSGHQLFALSRSQLERALGQDEGKRLYSQILVQRNVSGYKTTSASELQSILRKVREKVEVS
Summary
Uniprot
EMBL
NWSH01001113
PCG72549.1
PCG72548.1
PCG72551.1
KZ150258
PZC71771.1
+ More
KQ460077 KPJ17857.1 ODYU01009606 SOQ54156.1 GDRN01088665 JAI60817.1 GDRN01088666 JAI60816.1 GDRN01088661 JAI60820.1 GL732595 EFX72825.1 GDIQ01254594 JAJ97130.1 GDIQ01209003 JAK42722.1 GDIP01055995 JAM47720.1 GDIQ01016410 JAN78327.1 GDIP01150881 LRGB01002722 JAJ72521.1 KZS06454.1 GDRN01088664 GDRN01088663 JAI60818.1 MNPL01002588 OQR78153.1
KQ460077 KPJ17857.1 ODYU01009606 SOQ54156.1 GDRN01088665 JAI60817.1 GDRN01088666 JAI60816.1 GDRN01088661 JAI60820.1 GL732595 EFX72825.1 GDIQ01254594 JAJ97130.1 GDIQ01209003 JAK42722.1 GDIP01055995 JAM47720.1 GDIQ01016410 JAN78327.1 GDIP01150881 LRGB01002722 JAJ72521.1 KZS06454.1 GDRN01088664 GDRN01088663 JAI60818.1 MNPL01002588 OQR78153.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
PDB
1WXB
E-value=1.20268e-07,
Score=136
Ontologies
GO
PANTHER
Topology
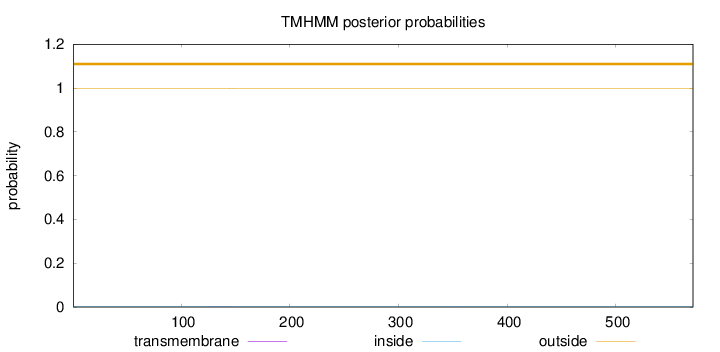
Length:
571
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00644
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00180
outside
1 - 571
Population Genetic Test Statistics
Pi
359.008973
Theta
181.316389
Tajima's D
2.773282
CLR
0.234532
CSRT
0.966351682415879
Interpretation
Uncertain
