Gene
KWMTBOMO16017
Pre Gene Modal
BGIBMGA004600
Annotation
PREDICTED:_IQ_motif_and_SEC7_domain-containing_protein_1_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.35
Sequence
CDS
ATGGAGAATCGTTGTCTTTGCTTACTTTCTCCGAACGATCGTGACTCCGGCAGTGGTTATGGTCTGGTTATGTGCTTTTCGCAGAAGGTAGACGAAGAGGAGCCAGGCGGCGGGGTTCCGCTGCATCCACCGGACGGCTTCTACACTCATCAGCACTCGATCCACGCGCATCATGCCCACACTGAGCCCGACCGAGGGTACATGGATGGCAACAGATACGTCACCGATGTCCATCGACCGTTACCAGCTCTAAATCATTCGAACTCGTCATGTTCGTCGGCGTCGAACGCGTCGACGGGCGCGGGTCTTTGCACGGCACAGGAGCCGGCGTCGCAGCTTTACATTGCCGCGCCGCCGGAGGGCTTCGGCCTCGCCGTTCATACCAACAGCCGCTATGAGATGTCGCAGGACCTGCTCGATAAGCAAGTGGAAATCTTAGAGAGACGATACGGAGGTCTCCGCGCCCGCAGGGCTGCCGTCACCATCCAGCGCGCGTTCCGAAGGCGGCAACTGCTCAAGAAGTTCAGCGCGATCACGGCTATGGCCAAAGCTGCCGACTCGAATGCGAGACGGCTACAAGACGGTCCCGATCACGGTCAATTAGTCAATGGTACTGATATCTACGCGACTACGTTGCTACAAAAAAGTCCCAGCGGCACAGAAAAGATGTTAGCTTTGACGAGACCCATGAGGTCGATGTCCTTGCGGGAGCGACACGAACCACCCGACATGGGCAACCACTTGCAATGCGACACGCACGACGTAGAAGCCATGGTTGCTAGAGTTCAGCAGTCGGCTCACCAGATCCACATGATCGCCACCGATCTACCGCCCGGGGTGCTTCATCCGGCTGACGCAGACAGCGGGGTTTGCAGTTGTTCAGTTAGCGGCTCCGTGACCAATATATCGACTTCATCATCGCACAATGAATCTCTACACAATCACCAGATGATGAATACTGGTCCGATCTATGGAAATCTACTCTCGTCGGGTGGCAAACCCCTGTCGTCTCCCACCCCCCGACAGCAGCAGCAACTGGCTGCGATACAAGCTGCGAACGCCAGGAGGATGCCTCCGGAAGTGCCCAAAAGAACGAGCAGCATAACTCTAAACGCCGAGAGTCCGGTGTCGCTTAGACGTGCGGAATGCGGAGCGGTCGTCAGGGGAGGATCGATGTCTTCCGTGCAGTCCTCGTCCTCATCATCATCGCAGGAGCATCGCCATCACTACCACCAGCCCTACCAGCCCAGGCAGCCGGAGACGCACGTGCACGTCCCAGAGTACCTCAACGCGCGTCCCGGCGTCGTGCTGGAAGGAGATCGGTACTCCGTCTGGAAGAGACAGGAGGCCGCTCCTTACTACGAAGCCCCCCCGCCCCCGATGGCAGCGCCCTGCTGTAGGCCACAGCATGAGCACCATCAGCATCAGCAACCGTTAAAGGTGTCAGAAACTGTCCGGAAACGCCAGTACCGCGTCGGCCTTAACTTGTTCAACAAGAAACCAGAGCGCGGCATCGCTTACCTGATCTCCCGCGGCTTCCTCGAGAACTCGCCGCGGGGCGTGGCAAGGTTCCTCATCACCAGAAAGGGCCTGAGCAAACAGATGATCGGAGAATACCTTGGAAACCTGCAGAATCCATTCAATATGGCAGTTCTGGAATGCTTCGTAAACGAGCTAGACTTGTCGGGTATGGCGGTAGACGTGGCTCTGCGCCGGTACCAGGCTCACTTCCGGTTGCCGGGCGAGGCGCAGAAGATCGAGCGTCTGGTCGAAGCGTTCGCCAGAAGATACTGCATCTGCAACACAGATTTCGTTCAAAGGTTGCGGACTCAGGACACGATATTCGTGTTGGCCTTCGCCATAATAATGCTGAACACGGATTTGCACACTCCGAACCTGAAGCCGGAAGCTCGCATGTCCTTGGAAGACTTCGTGAAGAACCTTCGCGGCATCGACGACTGCGGCGACATCGACAGGGACATGCTCGCTGGGATCTACGACAGAGTCAAGGCGAACGAGTTTAGACCCGGCAGCGACCACGTCACGCAGGTGATGAAAGTGCAAGCGACGATAGTCGGGAAGAAGCCCCACCTCGCTCTGCCGCACCGTCGCCTCGTCTGCTACTGCAGGTTGTACGAGATACCTGACATACACAAGAAGGAGCGACCCGGAGTCCATCAGAGAGAAGTATTCCTTTTCAACGATCTACTCGTGATCACGAAAATCTTCAGCAAGAAGAAATCGTCGGTGACGTACACCTTCCGACAGTCGTTTCCGCTGTGCGGCATGGTGGTCAGCCTGTTCTCCGTGCCTCATTACCCGTTCGGCATACGTTTGTCTCGTCGAGTCGACAACCGTCGCTTGTCTACGTTTAACGCTCGCAACGAGCACGACCGCTGCAAGTTCGCGGAGGACCTTCGCGAGAGTATCGCCGAGATGGACGAGATGGAGGCCATGAGGATCGAGGGCGAACTCAACAGGGGGAAGGGAAGGAACACCGGGAACAACGCCAGGAACGTGATGGAAAACCGAGACAGCGGCGTGGCTGATGTTGAAATCGATCACCAGCAGTGCGTCGACCACGTCCACATGCACGGCGTGGTGGTTCCTCCTCCGCCCTCGTGTCCAGCACCAGCAGCCCCCTCCAGGCTCTCCTCGAACCCACCGCCCACCAGCATCATCAAACGGAACGTGCTCAGCAACTCTCTAGTCGATATCAATGACCCGGCGGAGCGACTCCAGCGGCGCGGGTCGGTGGGGTCCCTGGACAGCGGCATGTCCGTCTCCTTCCAGCAGGGAAGCAGGAACGCATTACACGCCACTTCACCGAGACATCCCACGGAACAACGGTCGCCGGGTCGTTTGTTCGGCGGCATATTCCCGGGACGTATTCGTAAGCTCAGCGCCTCCGGGATCCCGGACGCGAAGCACGGGCAGGTCGGGAAGAGCACCGAAGTTTGA
Protein
MENRCLCLLSPNDRDSGSGYGLVMCFSQKVDEEEPGGGVPLHPPDGFYTHQHSIHAHHAHTEPDRGYMDGNRYVTDVHRPLPALNHSNSSCSSASNASTGAGLCTAQEPASQLYIAAPPEGFGLAVHTNSRYEMSQDLLDKQVEILERRYGGLRARRAAVTIQRAFRRRQLLKKFSAITAMAKAADSNARRLQDGPDHGQLVNGTDIYATTLLQKSPSGTEKMLALTRPMRSMSLRERHEPPDMGNHLQCDTHDVEAMVARVQQSAHQIHMIATDLPPGVLHPADADSGVCSCSVSGSVTNISTSSSHNESLHNHQMMNTGPIYGNLLSSGGKPLSSPTPRQQQQLAAIQAANARRMPPEVPKRTSSITLNAESPVSLRRAECGAVVRGGSMSSVQSSSSSSSQEHRHHYHQPYQPRQPETHVHVPEYLNARPGVVLEGDRYSVWKRQEAAPYYEAPPPPMAAPCCRPQHEHHQHQQPLKVSETVRKRQYRVGLNLFNKKPERGIAYLISRGFLENSPRGVARFLITRKGLSKQMIGEYLGNLQNPFNMAVLECFVNELDLSGMAVDVALRRYQAHFRLPGEAQKIERLVEAFARRYCICNTDFVQRLRTQDTIFVLAFAIIMLNTDLHTPNLKPEARMSLEDFVKNLRGIDDCGDIDRDMLAGIYDRVKANEFRPGSDHVTQVMKVQATIVGKKPHLALPHRRLVCYCRLYEIPDIHKKERPGVHQREVFLFNDLLVITKIFSKKKSSVTYTFRQSFPLCGMVVSLFSVPHYPFGIRLSRRVDNRRLSTFNARNEHDRCKFAEDLRESIAEMDEMEAMRIEGELNRGKGRNTGNNARNVMENRDSGVADVEIDHQQCVDHVHMHGVVVPPPPSCPAPAAPSRLSSNPPPTSIIKRNVLSNSLVDINDPAERLQRRGSVGSLDSGMSVSFQQGSRNALHATSPRHPTEQRSPGRLFGGIFPGRIRKLSASGIPDAKHGQVGKSTEV
Summary
Uniprot
EMBL
Proteomes
Interpro
SUPFAM
SSF48425
SSF48425
Gene 3D
ProteinModelPortal
PDB
6FNE
E-value=5.50838e-121,
Score=1115
Ontologies
GO
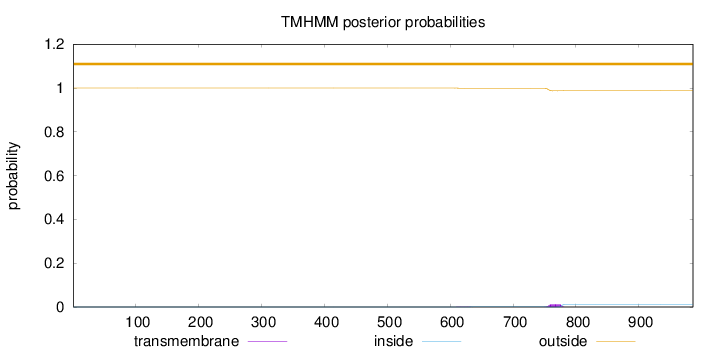
Topology
Length:
986
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.28935
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00011
outside
1 - 986
Population Genetic Test Statistics
Pi
278.017143
Theta
177.382733
Tajima's D
1.963262
CLR
0.140723
CSRT
0.876906154692265
Interpretation
Uncertain
