Gene
KWMTBOMO16004 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004541
Annotation
heat_shock_protein_20.4_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.106 Mitochondrial Reliability : 1.507
Sequence
CDS
ATGTCGTTGCTACCATATTTCTTCGATGATTTTGGCTCTAGACGCCCTCGCCGTCTGTTGGACCAGCATTTTGGCCTTGCCTTAACACCAGATGATCTTCTTAGTGTCGCTGCCGGGCCTTTGCTGAACAGGGAATACTACAGACCGTGGCGCCATCTCGCTGCAGCGGCTAGAGATGTTGGTTCCAGCATCAAAGTCGACAAGGATAAGTTCCAAGTGAATTTAGATGTGCAGCACTTCGCGCCAGAAGAGATCTCTGTGAAGACTGCCGACGGGTACATCGTAGTGGAAGGCAAGCACGAAGAGAAGAAAGACGAGCACGGGTATATTTCAAGGCAGTTCGTCCGACGTTACGCGCTGCCTGAAGGCGCGGCGCCTGAGACTGTGGAATCGCGACTGTCATCAGACGGGGTACTCACCATCACTGCGCCGAGGAAGGTGCCTGACGCCGTCAAGGGAGAGAGAAAGGTGCCCATCGCACAGACCGGTCCCGTTCGCAAGGAGATCAAGGATCAGAGTGAAGAAGCCAACGAGAAGGAAAAGTAG
Protein
MSLLPYFFDDFGSRRPRRLLDQHFGLALTPDDLLSVAAGPLLNREYYRPWRHLAAAARDVGSSIKVDKDKFQVNLDVQHFAPEEISVKTADGYIVVEGKHEEKKDEHGYISRQFVRRYALPEGAAPETVESRLSSDGVLTITAPRKVPDAVKGERKVPIAQTGPVRKEIKDQSEEANEKEK
Summary
Similarity
Belongs to the small heat shock protein (HSP20) family.
Uniprot
Q9GSB6
B9VTS8
A0A2H1W1Y8
E2F399
A0A2W1BHM1
A0A2A4J9F7
+ More
A0A0N1INR0 A0A0A7RE39 M1FKQ7 A0A0K0XQX1 A0A212EIS5 Q2LCS7 Q5MGN8 Q5MGL1 Q1HAT5 A0A161GJ22 Q5R1P7 E2F3A1 A0A2H1X0S0 A0A0C5PKY8 Q9GN07 A0A0P0CJ81 A0A0N0PBJ9 A0A3S2PJT0 Q0KKB1 S4P4G2 I4DMV4 A0A343EU23 A0A0A7BZM3 A0A0L7K2K7 A0A194QB65 A0A2H4LHL7 A0A2H4LI82 A0A2A4J3F8 A0A0A7BZF0 A0A2D1CRQ9 A0A2A4J9J4 A0A222NUK8 E2F397 A0A0L7L9D3 A0A212FJB9 A0A0A7BZ27 R4HL93 A0A0A7RCH5 O18634 A0A0A7RCE5 A0A220W0C8 A0A194Q6N8 A0A0N1PFQ5 A0A3S2L9M7 B2ZHW3 A0A2H1W2W7 F1C928 A0A194Q5N9 A0A385PHG6 A0A212F0A4 A0A0K0XQY0 A0A222NUL2 Q1AMF5 A0A212ERQ7 A0A212FIM4 D9J2L8 Q1AMF7 A0A1E1W5U5 A0A0A7BZ70 R9S0D1 A0A222NUL9 A0A212EIS8 Q5R1P6 B9VTS0 S4NMP5 R9RZX9 A0A2H1W170 A0A2A4JK93 A0A075BR54 A0A2A4JUN7 A0A2W1BH28 A0A2H1WH29 A0A222NUM2 F1C929 A0A2A4K9P8 A0A2D1CRP7 A0A2H1WKW0 E2F398 Q1AMF6 A0A2A4JLW2 A0A0K0XR46 A0A345W859 H9J540 A0A0N0PBK3 A0A194PVM7 A0A0L7L8L1 Q5R1P4 H9J4S5 A0A212FBC6 Q0KKB2 F1C930 A0A222NUK4 A0A212F079 A0A2W1BPZ5
A0A0N1INR0 A0A0A7RE39 M1FKQ7 A0A0K0XQX1 A0A212EIS5 Q2LCS7 Q5MGN8 Q5MGL1 Q1HAT5 A0A161GJ22 Q5R1P7 E2F3A1 A0A2H1X0S0 A0A0C5PKY8 Q9GN07 A0A0P0CJ81 A0A0N0PBJ9 A0A3S2PJT0 Q0KKB1 S4P4G2 I4DMV4 A0A343EU23 A0A0A7BZM3 A0A0L7K2K7 A0A194QB65 A0A2H4LHL7 A0A2H4LI82 A0A2A4J3F8 A0A0A7BZF0 A0A2D1CRQ9 A0A2A4J9J4 A0A222NUK8 E2F397 A0A0L7L9D3 A0A212FJB9 A0A0A7BZ27 R4HL93 A0A0A7RCH5 O18634 A0A0A7RCE5 A0A220W0C8 A0A194Q6N8 A0A0N1PFQ5 A0A3S2L9M7 B2ZHW3 A0A2H1W2W7 F1C928 A0A194Q5N9 A0A385PHG6 A0A212F0A4 A0A0K0XQY0 A0A222NUL2 Q1AMF5 A0A212ERQ7 A0A212FIM4 D9J2L8 Q1AMF7 A0A1E1W5U5 A0A0A7BZ70 R9S0D1 A0A222NUL9 A0A212EIS8 Q5R1P6 B9VTS0 S4NMP5 R9RZX9 A0A2H1W170 A0A2A4JK93 A0A075BR54 A0A2A4JUN7 A0A2W1BH28 A0A2H1WH29 A0A222NUM2 F1C929 A0A2A4K9P8 A0A2D1CRP7 A0A2H1WKW0 E2F398 Q1AMF6 A0A2A4JLW2 A0A0K0XR46 A0A345W859 H9J540 A0A0N0PBK3 A0A194PVM7 A0A0L7L8L1 Q5R1P4 H9J4S5 A0A212FBC6 Q0KKB2 F1C930 A0A222NUK4 A0A212F079 A0A2W1BPZ5
Pubmed
EMBL
BABH01025846
AF315318
AAG30945.2
FJ602788
ACM24354.1
ODYU01005718
+ More
SOQ46842.1 HM046615 ADK55522.1 KZ150055 PZC74338.1 NWSH01002265 PCG68757.1 KQ460891 KPJ10837.1 KM217077 KM217078 AJA32865.1 JQ708200 AFQ02692.1 KP843896 AKS40075.1 AGBW02014566 OWR41396.1 DQ336356 ABC68342.2 AY829748 AAV91362.1 AY829747 AY829775 AAV91361.1 AAV91389.1 JX491641 KC710020 AB244517 AGC23337.1 AGM90553.1 BAE94664.1 KU096844 AMY63177.1 AB195970 BAD74195.1 HM046617 ADK55524.1 ODYU01012514 SOQ58867.1 KM881570 AJQ22645.1 AF315317 AF315319 FJ602772 AAG30944.1 AAG30946.1 ACM24338.1 KR137569 ALI87024.1 KPJ10841.1 RSAL01000012 RVE53492.1 AB251898 BAF03558.1 GAIX01009047 JAA83513.1 AK402622 BAM19244.1 MF067296 ASM61894.1 KJ461917 AHW45919.1 JTDY01014370 KOB52036.1 KQ459460 KPJ00666.1 KX845562 ATB54993.1 PZC74337.1 KX845559 ATB54990.1 PZC74333.1 NWSH01003339 PCG66520.1 PCG66521.1 KJ461916 AHW45918.1 KX977584 ATN45243.1 PCG68755.1 KX958476 ASQ43192.1 HM046613 ADK55520.1 JTDY01002160 KOB71979.1 AGBW02008280 OWR53838.1 KJ461920 AHW45922.1 HQ647012 AET10424.1 KM217079 AJA32867.1 U94328 AAC36146.1 KM217080 AJA32868.1 KY701309 ASK86231.1 KPJ00670.1 KPJ10838.1 RSAL01000078 RVE48718.1 EU668902 ACD01216.1 SOQ46844.1 HQ219475 ADX96000.1 KPJ00669.1 MG816464 AYA93248.1 AGBW02011158 OWR47131.1 KP843906 AKS40085.1 KX958479 ASQ43195.1 DQ063591 AAZ14792.1 AGBW02012988 OWR44131.1 AGBW02008361 OWR53574.1 HM359007 ADI48314.1 DQ063589 AAZ14790.1 GDQN01008714 JAT82340.1 KJ461913 AHW45915.1 KC710018 KC710022 AGM90551.1 AGM90555.1 KX958482 ASQ43198.1 OWR41397.1 AB195971 BAD74196.1 FJ602780 ACM24346.1 GAIX01014246 JAA78314.1 KC710019 KC710024 AGM90552.1 AGM90557.1 RVE53491.1 SOQ46845.1 NWSH01001148 PCG72391.1 KC689795 KX845561 KZ150027 AGQ42773.1 ATB54992.1 PZC74791.1 NWSH01000548 PCG75721.1 KZ150133 PZC73025.1 ODYU01008627 SOQ52368.1 KX958478 ASQ43194.1 HQ219476 ADX96001.1 NWSH01000008 PCG80951.1 KX977585 ATN45244.1 ODYU01009326 SOQ53668.1 HM046614 ADK55521.1 DQ063590 AAZ14791.1 PCG72390.1 KP843898 AKS40077.1 MH042022 AXJ21597.1 BABH01042227 KQ460970 KPJ10176.1 KQ459597 KPI95180.1 JTDY01002346 KOB71621.1 AB195973 BAD74198.1 BABH01035507 AGBW02009356 OWR51030.1 AB251897 BAF03557.1 HQ219477 ADX96002.1 KX958471 ASQ43187.1 OWR47132.1 PZC74790.1
SOQ46842.1 HM046615 ADK55522.1 KZ150055 PZC74338.1 NWSH01002265 PCG68757.1 KQ460891 KPJ10837.1 KM217077 KM217078 AJA32865.1 JQ708200 AFQ02692.1 KP843896 AKS40075.1 AGBW02014566 OWR41396.1 DQ336356 ABC68342.2 AY829748 AAV91362.1 AY829747 AY829775 AAV91361.1 AAV91389.1 JX491641 KC710020 AB244517 AGC23337.1 AGM90553.1 BAE94664.1 KU096844 AMY63177.1 AB195970 BAD74195.1 HM046617 ADK55524.1 ODYU01012514 SOQ58867.1 KM881570 AJQ22645.1 AF315317 AF315319 FJ602772 AAG30944.1 AAG30946.1 ACM24338.1 KR137569 ALI87024.1 KPJ10841.1 RSAL01000012 RVE53492.1 AB251898 BAF03558.1 GAIX01009047 JAA83513.1 AK402622 BAM19244.1 MF067296 ASM61894.1 KJ461917 AHW45919.1 JTDY01014370 KOB52036.1 KQ459460 KPJ00666.1 KX845562 ATB54993.1 PZC74337.1 KX845559 ATB54990.1 PZC74333.1 NWSH01003339 PCG66520.1 PCG66521.1 KJ461916 AHW45918.1 KX977584 ATN45243.1 PCG68755.1 KX958476 ASQ43192.1 HM046613 ADK55520.1 JTDY01002160 KOB71979.1 AGBW02008280 OWR53838.1 KJ461920 AHW45922.1 HQ647012 AET10424.1 KM217079 AJA32867.1 U94328 AAC36146.1 KM217080 AJA32868.1 KY701309 ASK86231.1 KPJ00670.1 KPJ10838.1 RSAL01000078 RVE48718.1 EU668902 ACD01216.1 SOQ46844.1 HQ219475 ADX96000.1 KPJ00669.1 MG816464 AYA93248.1 AGBW02011158 OWR47131.1 KP843906 AKS40085.1 KX958479 ASQ43195.1 DQ063591 AAZ14792.1 AGBW02012988 OWR44131.1 AGBW02008361 OWR53574.1 HM359007 ADI48314.1 DQ063589 AAZ14790.1 GDQN01008714 JAT82340.1 KJ461913 AHW45915.1 KC710018 KC710022 AGM90551.1 AGM90555.1 KX958482 ASQ43198.1 OWR41397.1 AB195971 BAD74196.1 FJ602780 ACM24346.1 GAIX01014246 JAA78314.1 KC710019 KC710024 AGM90552.1 AGM90557.1 RVE53491.1 SOQ46845.1 NWSH01001148 PCG72391.1 KC689795 KX845561 KZ150027 AGQ42773.1 ATB54992.1 PZC74791.1 NWSH01000548 PCG75721.1 KZ150133 PZC73025.1 ODYU01008627 SOQ52368.1 KX958478 ASQ43194.1 HQ219476 ADX96001.1 NWSH01000008 PCG80951.1 KX977585 ATN45244.1 ODYU01009326 SOQ53668.1 HM046614 ADK55521.1 DQ063590 AAZ14791.1 PCG72390.1 KP843898 AKS40077.1 MH042022 AXJ21597.1 BABH01042227 KQ460970 KPJ10176.1 KQ459597 KPI95180.1 JTDY01002346 KOB71621.1 AB195973 BAD74198.1 BABH01035507 AGBW02009356 OWR51030.1 AB251897 BAF03557.1 HQ219477 ADX96002.1 KX958471 ASQ43187.1 OWR47132.1 PZC74790.1
Proteomes
Pfam
PF00011 HSP20
Interpro
Gene 3D
CDD
ProteinModelPortal
Q9GSB6
B9VTS8
A0A2H1W1Y8
E2F399
A0A2W1BHM1
A0A2A4J9F7
+ More
A0A0N1INR0 A0A0A7RE39 M1FKQ7 A0A0K0XQX1 A0A212EIS5 Q2LCS7 Q5MGN8 Q5MGL1 Q1HAT5 A0A161GJ22 Q5R1P7 E2F3A1 A0A2H1X0S0 A0A0C5PKY8 Q9GN07 A0A0P0CJ81 A0A0N0PBJ9 A0A3S2PJT0 Q0KKB1 S4P4G2 I4DMV4 A0A343EU23 A0A0A7BZM3 A0A0L7K2K7 A0A194QB65 A0A2H4LHL7 A0A2H4LI82 A0A2A4J3F8 A0A0A7BZF0 A0A2D1CRQ9 A0A2A4J9J4 A0A222NUK8 E2F397 A0A0L7L9D3 A0A212FJB9 A0A0A7BZ27 R4HL93 A0A0A7RCH5 O18634 A0A0A7RCE5 A0A220W0C8 A0A194Q6N8 A0A0N1PFQ5 A0A3S2L9M7 B2ZHW3 A0A2H1W2W7 F1C928 A0A194Q5N9 A0A385PHG6 A0A212F0A4 A0A0K0XQY0 A0A222NUL2 Q1AMF5 A0A212ERQ7 A0A212FIM4 D9J2L8 Q1AMF7 A0A1E1W5U5 A0A0A7BZ70 R9S0D1 A0A222NUL9 A0A212EIS8 Q5R1P6 B9VTS0 S4NMP5 R9RZX9 A0A2H1W170 A0A2A4JK93 A0A075BR54 A0A2A4JUN7 A0A2W1BH28 A0A2H1WH29 A0A222NUM2 F1C929 A0A2A4K9P8 A0A2D1CRP7 A0A2H1WKW0 E2F398 Q1AMF6 A0A2A4JLW2 A0A0K0XR46 A0A345W859 H9J540 A0A0N0PBK3 A0A194PVM7 A0A0L7L8L1 Q5R1P4 H9J4S5 A0A212FBC6 Q0KKB2 F1C930 A0A222NUK4 A0A212F079 A0A2W1BPZ5
A0A0N1INR0 A0A0A7RE39 M1FKQ7 A0A0K0XQX1 A0A212EIS5 Q2LCS7 Q5MGN8 Q5MGL1 Q1HAT5 A0A161GJ22 Q5R1P7 E2F3A1 A0A2H1X0S0 A0A0C5PKY8 Q9GN07 A0A0P0CJ81 A0A0N0PBJ9 A0A3S2PJT0 Q0KKB1 S4P4G2 I4DMV4 A0A343EU23 A0A0A7BZM3 A0A0L7K2K7 A0A194QB65 A0A2H4LHL7 A0A2H4LI82 A0A2A4J3F8 A0A0A7BZF0 A0A2D1CRQ9 A0A2A4J9J4 A0A222NUK8 E2F397 A0A0L7L9D3 A0A212FJB9 A0A0A7BZ27 R4HL93 A0A0A7RCH5 O18634 A0A0A7RCE5 A0A220W0C8 A0A194Q6N8 A0A0N1PFQ5 A0A3S2L9M7 B2ZHW3 A0A2H1W2W7 F1C928 A0A194Q5N9 A0A385PHG6 A0A212F0A4 A0A0K0XQY0 A0A222NUL2 Q1AMF5 A0A212ERQ7 A0A212FIM4 D9J2L8 Q1AMF7 A0A1E1W5U5 A0A0A7BZ70 R9S0D1 A0A222NUL9 A0A212EIS8 Q5R1P6 B9VTS0 S4NMP5 R9RZX9 A0A2H1W170 A0A2A4JK93 A0A075BR54 A0A2A4JUN7 A0A2W1BH28 A0A2H1WH29 A0A222NUM2 F1C929 A0A2A4K9P8 A0A2D1CRP7 A0A2H1WKW0 E2F398 Q1AMF6 A0A2A4JLW2 A0A0K0XR46 A0A345W859 H9J540 A0A0N0PBK3 A0A194PVM7 A0A0L7L8L1 Q5R1P4 H9J4S5 A0A212FBC6 Q0KKB2 F1C930 A0A222NUK4 A0A212F079 A0A2W1BPZ5
PDB
3J07
E-value=8.01095e-19,
Score=226
Ontologies
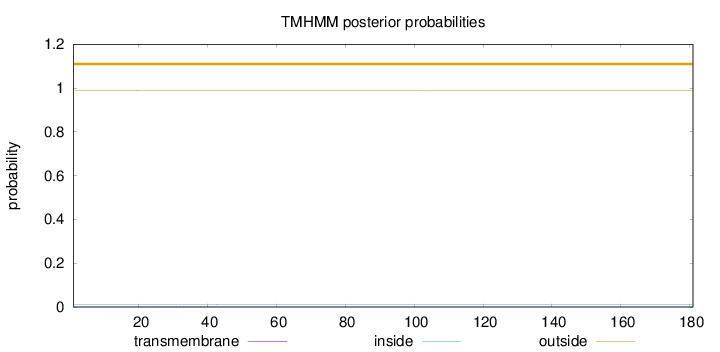
Topology
Length:
181
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0273
Exp number, first 60 AAs:
0.0273
Total prob of N-in:
0.01148
outside
1 - 181
Population Genetic Test Statistics
Pi
142.476914
Theta
195.135143
Tajima's D
-0.825782
CLR
0.07942
CSRT
0.173491325433728
Interpretation
Uncertain
