Gene
KWMTBOMO15997
Pre Gene Modal
BGIBMGA004611
Annotation
PREDICTED:_dual_specificity_tyrosine-phosphorylation-regulated_kinase_2-like_[Bombyx_mori]
Full name
Putative dual specificity tyrosine-phosphorylation-regulated kinase 3 homolog
+ More
Dual specificity tyrosine-phosphorylation-regulated kinase 2
Dual specificity tyrosine-phosphorylation-regulated kinase 2
Location in the cell
Mitochondrial Reliability : 1.515 Nuclear Reliability : 1.341
Sequence
CDS
ATGCCTCTGGGGAGCGCGCACGCGTTCGGGCTCGCGTTGCTCCGGGACGGGGCACTGCCGCCGCTGGAGCCTGGCCCGCCCGCACCGCCGGCAGCCAACGCGCAACCGCCCGACAAGCGACCGACGGCCGCCGCCGCCAGTCCCGAGCAGGTCATGAAGCTCTATATGAACAAACTCACGCCTTACGAGCACCGTGAAATATTCGACTATCCTCAAGTGTACTTCATAGGTGCGAACGCGAAGAAGCGCCCCGGCCTAGTCGGGTTCCCAAACAATTGCGAGTATGACAACGAGCAGGGCTCGTATATCCACATCCCCCACGACCACATAGCCTACAGATACGAAGTGCTGAAAGTCATCGGCAAAGGTAGCTTCGGGCAAGTGGTGAAAGCGTTCGATCATAAGAAACGCGAACACGTTGCCCTCAAGATGGTCCGCAACGAAAAGCGGTTTCACAGACAAGCACAGGAGGAGATCCGTATACTGGAGCACCTCCGCGAGCAAGACAAGGACAACACGATGAACGTCATCCACATGTTCGACTCGTTCACATTCAGGAATCACACGTGCATCACTTTCGAGCTACTCTCGATCAACTTGTACGAGTTGATAAAGAAGAACAAGTTCCAAGGTTTCTCTCTGCAGTTGGTGCGTAAGTTCTCGCATAGTCTTCTGCAGTGTCTGCACGCTCTAAACAAGAACCGCATCATCCACTGCGACATGAAGCCAGAGAACGTGCTGCTCAAGCAACAGGGACGATCTGGTATCAAGGTCAGTATGCTTAAAATGATTATAACCGGTTGTGTAAAAATGATACTACTATCTATAAAATACCGAGTAATAATCAAACCAATTGCAGTCCTTATGCATATTTCGAAATTTTCATTATTAGTGTTCCATTAG
Protein
MPLGSAHAFGLALLRDGALPPLEPGPPAPPAANAQPPDKRPTAAAASPEQVMKLYMNKLTPYEHREIFDYPQVYFIGANAKKRPGLVGFPNNCEYDNEQGSYIHIPHDHIAYRYEVLKVIGKGSFGQVVKAFDHKKREHVALKMVRNEKRFHRQAQEEIRILEHLREQDKDNTMNVIHMFDSFTFRNHTCITFELLSINLYELIKKNKFQGFSLQLVRKFSHSLLQCLHALNKNRIIHCDMKPENVLLKQQGRSGIKVSMLKMIITGCVKMILLSIKYRVIIKPIAVLMHISKFSLLVFH
Summary
Description
Serine/threonine-protein kinase involved in the control of mitotic transition and the regulation of cellular growth and/or development.
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Cofactor
Mg(2+)
Mn(2+)
Mn(2+)
Subunit
Interacts with MDM2.
Similarity
Belongs to the protein kinase superfamily.
Belongs to the protein kinase superfamily. CMGC Ser/Thr protein kinase family. MNB/DYRK subfamily.
Belongs to the protein kinase superfamily. CMGC Ser/Thr protein kinase family. MNB/DYRK subfamily.
Keywords
ATP-binding
Complete proteome
Kinase
Nucleotide-binding
Phosphoprotein
Reference proteome
Transferase
Tyrosine-protein kinase
Apoptosis
Cytoplasm
Magnesium
Manganese
Nucleus
Ubl conjugation
Feature
chain Putative dual specificity tyrosine-phosphorylation-regulated kinase 3 homolog
Uniprot
H9J521
A0A2A4J084
A0A2W1BNM3
A0A2H1WZK8
A0A222AJ63
A0A194Q549
+ More
A0A0N0PBK1 V5I7X1 A0A1Y1L2W6 A0A2J7PXY8 A0A2P8XTB8 A0A0T6B1H5 D6W776 E9IBY7 F4X1P3 A0A151WNG7 K7J5L1 E2ACJ1 A0A1B6CMA4 A0A232F437 A0A151IW63 A0A195BEK7 A0A195C9L9 E2BBP0 A0A2R5LMB7 A0A293LS30 A0A158NQU4 A0A195F5I2 A0A2A3ETQ0 T1JLC7 A0A088ALP9 A0A0M9A182 A0A1B6LKS8 E0W1C0 A0A1B6HJ39 A0A3L8DTD2 L7LW76 L7LT75 A0A131XUL6 A0A1A9UMB8 A0A1A9ZM30 A0A0P4WDQ9 A0A310S6M1 A0A087SVP6 N6T9F7 A0A099YYF6 A0A093JLF6 B4PW56 A0A0L0BL59 A0A0C9R8B8 A0A1A9WQC1 B3P9N5 A0A0J9S1A8 A0A1I8M0W0 P83102 B4IKS4 A0A0J9S1C0 T1PLQ1 U3KJU8 A0A091QHR0 A0A1A9YKB5 A0A1B0BID7 B4NHG7 W8C7X9 A0A2Y9E3X7 M1EJV5 A0A1V9X078 A0A212DF93 A0A2K6G6Z1 L8HRY6 A0A2K5SCX2 A0A3B0KXV9 F1RYI2 A0A3P8Y962 W5Q8S2 A0A3Q7SPL6 A0A286ZM94 H0XS18 A0A1B0FDU3 E1BG90 A0A1U7RM61 A0A3B4UQW6 T0MIS9 A0A3P8YBD9 U6DY67 A0A2K6SL42 M3Z5N1 A0A2K6SL48 A0A2U3X3H9 M3WEG8 Q5ZIU3 U3DTS3 A0A091EQ34 E2R3D8 A0A2P4SVP3 B4JZR2
A0A0N0PBK1 V5I7X1 A0A1Y1L2W6 A0A2J7PXY8 A0A2P8XTB8 A0A0T6B1H5 D6W776 E9IBY7 F4X1P3 A0A151WNG7 K7J5L1 E2ACJ1 A0A1B6CMA4 A0A232F437 A0A151IW63 A0A195BEK7 A0A195C9L9 E2BBP0 A0A2R5LMB7 A0A293LS30 A0A158NQU4 A0A195F5I2 A0A2A3ETQ0 T1JLC7 A0A088ALP9 A0A0M9A182 A0A1B6LKS8 E0W1C0 A0A1B6HJ39 A0A3L8DTD2 L7LW76 L7LT75 A0A131XUL6 A0A1A9UMB8 A0A1A9ZM30 A0A0P4WDQ9 A0A310S6M1 A0A087SVP6 N6T9F7 A0A099YYF6 A0A093JLF6 B4PW56 A0A0L0BL59 A0A0C9R8B8 A0A1A9WQC1 B3P9N5 A0A0J9S1A8 A0A1I8M0W0 P83102 B4IKS4 A0A0J9S1C0 T1PLQ1 U3KJU8 A0A091QHR0 A0A1A9YKB5 A0A1B0BID7 B4NHG7 W8C7X9 A0A2Y9E3X7 M1EJV5 A0A1V9X078 A0A212DF93 A0A2K6G6Z1 L8HRY6 A0A2K5SCX2 A0A3B0KXV9 F1RYI2 A0A3P8Y962 W5Q8S2 A0A3Q7SPL6 A0A286ZM94 H0XS18 A0A1B0FDU3 E1BG90 A0A1U7RM61 A0A3B4UQW6 T0MIS9 A0A3P8YBD9 U6DY67 A0A2K6SL42 M3Z5N1 A0A2K6SL48 A0A2U3X3H9 M3WEG8 Q5ZIU3 U3DTS3 A0A091EQ34 E2R3D8 A0A2P4SVP3 B4JZR2
EC Number
2.7.12.1
Pubmed
19121390
28756777
26354079
28004739
29403074
18362917
+ More
19820115 21282665 21719571 20075255 20798317 28648823 21347285 20566863 30249741 25576852 29652888 23537049 17994087 17550304 26108605 22936249 25315136 10731132 12537572 12537569 18327897 24495485 23236062 28327890 29294181 22751099 25069045 20809919 19393038 23149746 17975172 15642098 25243066 16341006
19820115 21282665 21719571 20075255 20798317 28648823 21347285 20566863 30249741 25576852 29652888 23537049 17994087 17550304 26108605 22936249 25315136 10731132 12537572 12537569 18327897 24495485 23236062 28327890 29294181 22751099 25069045 20809919 19393038 23149746 17975172 15642098 25243066 16341006
EMBL
BABH01025857
NWSH01004019
PCG65567.1
KZ150055
PZC74330.1
ODYU01012242
+ More
SOQ58447.1 KY609556 ASO76420.1 KQ459460 KPJ00663.1 KQ460891 KPJ10844.1 GALX01005695 JAB62771.1 GEZM01066361 JAV67924.1 NEVH01020852 PNF21201.1 PYGN01001379 PSN35261.1 LJIG01016290 KRT81044.1 KQ971307 EFA11400.2 GL762138 EFZ21916.1 GL888544 EGI59634.1 KQ982907 KYQ49383.1 GL438528 EFN68857.1 GEDC01022783 JAS14515.1 NNAY01001002 OXU25534.1 KQ980874 KYN12044.1 KQ976500 KYM83008.1 KQ978068 KYM97410.1 GL447128 EFN86917.1 GGLE01006544 MBY10670.1 GFWV01004910 MAA29640.1 ADTU01023607 ADTU01023608 ADTU01023609 ADTU01023610 KQ981810 KYN35437.1 KZ288189 PBC34586.1 JH430326 KQ435794 KOX73803.1 GEBQ01015687 JAT24290.1 DS235867 EEB19426.1 GECU01033019 JAS74687.1 QOIP01000004 RLU23705.1 GACK01008989 JAA56045.1 GACK01009743 JAA55291.1 GEFM01006450 GEGO01001631 JAP69346.1 JAR93773.1 GDRN01097294 JAI59149.1 KQ767803 OAD53297.1 KK112173 KFM56935.1 APGK01039173 KB740967 KB632399 ENN76889.1 ERL94622.1 KL886538 KGL73933.1 KL206175 KFV79604.1 CM000161 EDW99361.1 KRK05016.1 JRES01001702 KNC20801.1 GBYB01004258 GBYB01004260 JAG74025.1 JAG74027.1 CH954184 EDV45198.1 CM002916 KMZ07417.1 AE014135 AY118401 AF103941 CH480855 EDW52664.1 KMZ07418.1 KMZ07419.1 KMZ07420.1 KA648975 AFP63604.1 AGTO01020282 KK698728 KFQ25976.1 JXJN01014918 JXJN01014919 CH964272 EDW83536.2 GAMC01007776 JAB98779.1 JP009043 AER97640.1 MNPL01031106 OQR66796.1 MKHE01000003 OWK16927.1 JH883377 ELR46631.1 OUUW01000017 SPP88918.1 AEMK02000033 AMGL01081100 AAQR03139938 AAQR03139939 CCAG010003810 KB017114 EQB78420.1 HAAF01015576 CCP87396.1 AEYP01031577 AANG04003655 AJ720691 CAG32350.1 GAMT01003581 GAMS01006677 GAMR01007047 GAMQ01006234 JAB08280.1 JAB16459.1 JAB26885.1 JAB35617.1 KK718745 KFO58439.1 AAEX03007035 AAEX03007036 PPHD01020415 POI28195.1 CH916380 EDV90928.1
SOQ58447.1 KY609556 ASO76420.1 KQ459460 KPJ00663.1 KQ460891 KPJ10844.1 GALX01005695 JAB62771.1 GEZM01066361 JAV67924.1 NEVH01020852 PNF21201.1 PYGN01001379 PSN35261.1 LJIG01016290 KRT81044.1 KQ971307 EFA11400.2 GL762138 EFZ21916.1 GL888544 EGI59634.1 KQ982907 KYQ49383.1 GL438528 EFN68857.1 GEDC01022783 JAS14515.1 NNAY01001002 OXU25534.1 KQ980874 KYN12044.1 KQ976500 KYM83008.1 KQ978068 KYM97410.1 GL447128 EFN86917.1 GGLE01006544 MBY10670.1 GFWV01004910 MAA29640.1 ADTU01023607 ADTU01023608 ADTU01023609 ADTU01023610 KQ981810 KYN35437.1 KZ288189 PBC34586.1 JH430326 KQ435794 KOX73803.1 GEBQ01015687 JAT24290.1 DS235867 EEB19426.1 GECU01033019 JAS74687.1 QOIP01000004 RLU23705.1 GACK01008989 JAA56045.1 GACK01009743 JAA55291.1 GEFM01006450 GEGO01001631 JAP69346.1 JAR93773.1 GDRN01097294 JAI59149.1 KQ767803 OAD53297.1 KK112173 KFM56935.1 APGK01039173 KB740967 KB632399 ENN76889.1 ERL94622.1 KL886538 KGL73933.1 KL206175 KFV79604.1 CM000161 EDW99361.1 KRK05016.1 JRES01001702 KNC20801.1 GBYB01004258 GBYB01004260 JAG74025.1 JAG74027.1 CH954184 EDV45198.1 CM002916 KMZ07417.1 AE014135 AY118401 AF103941 CH480855 EDW52664.1 KMZ07418.1 KMZ07419.1 KMZ07420.1 KA648975 AFP63604.1 AGTO01020282 KK698728 KFQ25976.1 JXJN01014918 JXJN01014919 CH964272 EDW83536.2 GAMC01007776 JAB98779.1 JP009043 AER97640.1 MNPL01031106 OQR66796.1 MKHE01000003 OWK16927.1 JH883377 ELR46631.1 OUUW01000017 SPP88918.1 AEMK02000033 AMGL01081100 AAQR03139938 AAQR03139939 CCAG010003810 KB017114 EQB78420.1 HAAF01015576 CCP87396.1 AEYP01031577 AANG04003655 AJ720691 CAG32350.1 GAMT01003581 GAMS01006677 GAMR01007047 GAMQ01006234 JAB08280.1 JAB16459.1 JAB26885.1 JAB35617.1 KK718745 KFO58439.1 AAEX03007035 AAEX03007036 PPHD01020415 POI28195.1 CH916380 EDV90928.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000235965
UP000245037
+ More
UP000007266 UP000007755 UP000075809 UP000002358 UP000000311 UP000215335 UP000078492 UP000078540 UP000078542 UP000008237 UP000005205 UP000078541 UP000242457 UP000005203 UP000053105 UP000009046 UP000279307 UP000078200 UP000092445 UP000054359 UP000019118 UP000030742 UP000053641 UP000053584 UP000002282 UP000037069 UP000091820 UP000008711 UP000095301 UP000000803 UP000001292 UP000016665 UP000092443 UP000092460 UP000007798 UP000248480 UP000192247 UP000233160 UP000233040 UP000268350 UP000008227 UP000265140 UP000002356 UP000286640 UP000005225 UP000092444 UP000009136 UP000189705 UP000261420 UP000233220 UP000000715 UP000245340 UP000011712 UP000000539 UP000008225 UP000052976 UP000002254 UP000001070
UP000007266 UP000007755 UP000075809 UP000002358 UP000000311 UP000215335 UP000078492 UP000078540 UP000078542 UP000008237 UP000005205 UP000078541 UP000242457 UP000005203 UP000053105 UP000009046 UP000279307 UP000078200 UP000092445 UP000054359 UP000019118 UP000030742 UP000053641 UP000053584 UP000002282 UP000037069 UP000091820 UP000008711 UP000095301 UP000000803 UP000001292 UP000016665 UP000092443 UP000092460 UP000007798 UP000248480 UP000192247 UP000233160 UP000233040 UP000268350 UP000008227 UP000265140 UP000002356 UP000286640 UP000005225 UP000092444 UP000009136 UP000189705 UP000261420 UP000233220 UP000000715 UP000245340 UP000011712 UP000000539 UP000008225 UP000052976 UP000002254 UP000001070
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H9J521
A0A2A4J084
A0A2W1BNM3
A0A2H1WZK8
A0A222AJ63
A0A194Q549
+ More
A0A0N0PBK1 V5I7X1 A0A1Y1L2W6 A0A2J7PXY8 A0A2P8XTB8 A0A0T6B1H5 D6W776 E9IBY7 F4X1P3 A0A151WNG7 K7J5L1 E2ACJ1 A0A1B6CMA4 A0A232F437 A0A151IW63 A0A195BEK7 A0A195C9L9 E2BBP0 A0A2R5LMB7 A0A293LS30 A0A158NQU4 A0A195F5I2 A0A2A3ETQ0 T1JLC7 A0A088ALP9 A0A0M9A182 A0A1B6LKS8 E0W1C0 A0A1B6HJ39 A0A3L8DTD2 L7LW76 L7LT75 A0A131XUL6 A0A1A9UMB8 A0A1A9ZM30 A0A0P4WDQ9 A0A310S6M1 A0A087SVP6 N6T9F7 A0A099YYF6 A0A093JLF6 B4PW56 A0A0L0BL59 A0A0C9R8B8 A0A1A9WQC1 B3P9N5 A0A0J9S1A8 A0A1I8M0W0 P83102 B4IKS4 A0A0J9S1C0 T1PLQ1 U3KJU8 A0A091QHR0 A0A1A9YKB5 A0A1B0BID7 B4NHG7 W8C7X9 A0A2Y9E3X7 M1EJV5 A0A1V9X078 A0A212DF93 A0A2K6G6Z1 L8HRY6 A0A2K5SCX2 A0A3B0KXV9 F1RYI2 A0A3P8Y962 W5Q8S2 A0A3Q7SPL6 A0A286ZM94 H0XS18 A0A1B0FDU3 E1BG90 A0A1U7RM61 A0A3B4UQW6 T0MIS9 A0A3P8YBD9 U6DY67 A0A2K6SL42 M3Z5N1 A0A2K6SL48 A0A2U3X3H9 M3WEG8 Q5ZIU3 U3DTS3 A0A091EQ34 E2R3D8 A0A2P4SVP3 B4JZR2
A0A0N0PBK1 V5I7X1 A0A1Y1L2W6 A0A2J7PXY8 A0A2P8XTB8 A0A0T6B1H5 D6W776 E9IBY7 F4X1P3 A0A151WNG7 K7J5L1 E2ACJ1 A0A1B6CMA4 A0A232F437 A0A151IW63 A0A195BEK7 A0A195C9L9 E2BBP0 A0A2R5LMB7 A0A293LS30 A0A158NQU4 A0A195F5I2 A0A2A3ETQ0 T1JLC7 A0A088ALP9 A0A0M9A182 A0A1B6LKS8 E0W1C0 A0A1B6HJ39 A0A3L8DTD2 L7LW76 L7LT75 A0A131XUL6 A0A1A9UMB8 A0A1A9ZM30 A0A0P4WDQ9 A0A310S6M1 A0A087SVP6 N6T9F7 A0A099YYF6 A0A093JLF6 B4PW56 A0A0L0BL59 A0A0C9R8B8 A0A1A9WQC1 B3P9N5 A0A0J9S1A8 A0A1I8M0W0 P83102 B4IKS4 A0A0J9S1C0 T1PLQ1 U3KJU8 A0A091QHR0 A0A1A9YKB5 A0A1B0BID7 B4NHG7 W8C7X9 A0A2Y9E3X7 M1EJV5 A0A1V9X078 A0A212DF93 A0A2K6G6Z1 L8HRY6 A0A2K5SCX2 A0A3B0KXV9 F1RYI2 A0A3P8Y962 W5Q8S2 A0A3Q7SPL6 A0A286ZM94 H0XS18 A0A1B0FDU3 E1BG90 A0A1U7RM61 A0A3B4UQW6 T0MIS9 A0A3P8YBD9 U6DY67 A0A2K6SL42 M3Z5N1 A0A2K6SL48 A0A2U3X3H9 M3WEG8 Q5ZIU3 U3DTS3 A0A091EQ34 E2R3D8 A0A2P4SVP3 B4JZR2
PDB
5ZTN
E-value=7.40634e-103,
Score=954
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
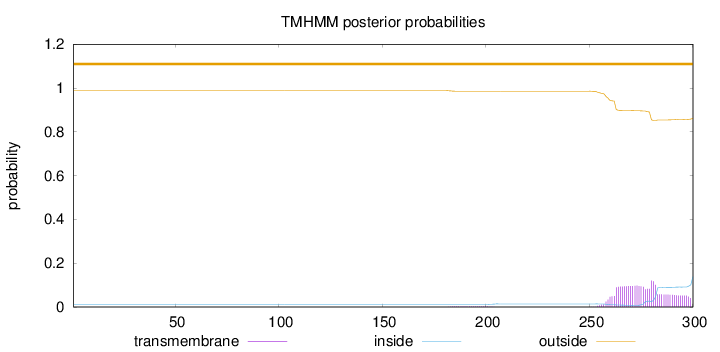
Length:
300
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.11674
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01142
outside
1 - 300
Population Genetic Test Statistics
Pi
215.768392
Theta
231.606095
Tajima's D
-0.197796
CLR
465.869401
CSRT
0.313684315784211
Interpretation
Uncertain
