Gene
KWMTBOMO15994
Annotation
PREDICTED:_dual_specificity_tyrosine-phosphorylation-regulated_kinase_2_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 1.294 PlasmaMembrane Reliability : 1.007
Sequence
CDS
ATGCCGCCACCCACCTTGAAACATAACTTCCAAGGTCTCACTTTTAATTTAATGAACGCTTTTATTTCCTTCCAGGTGATAGACTTCGGCAGCTCTTGCTATGAGCACCAACGCGTCTACACGTACATACAGTCGCGGTTCTATCGCGCGCCTGAGGTGATGATGGGCGCCCGATACGGTATGCCCATCGACATGTGGTCCCTTGGGTGCATACTCGCGGAGTTACTCATTGGGTTCCCCTTACTACCCGGCGAAGACGAAGCGGACCAAATGGCATGCATCATAGAGCTGCTAGGCATGCCTCCCCCCAAGTTAATAGAACAGGGGAAACGGTCAAAGAACTTCATCAGCAGCAAGGGCCTGCCGCGATACTGCACGGCAACCACCCTCGCCGACGGAACCACCGTGCTCAGTGGAGGCATGTCGAGGAGAGGCAAGCCCCGAGGTCCGCCGGGATCGAAGAGTTTTGTTACTGCACTAAAAGGATGTCAAGACAAATTTTTCATTGATTTCATTAGACGTTGCTTAGAATGGGACCCAGAGAAGCGCCTCACGCCGGCCGCAGCGCTACGCCATGCGTGGCTTCGGAGACGGCTGCCGAGGGCGCCCCACGACGACGAACCCAACCACGTTAGTATCGCGCACCACGCCAAGCGCCTCGCGCACCCCAACTGCACTGACAAATAG
Protein
MPPPTLKHNFQGLTFNLMNAFISFQVIDFGSSCYEHQRVYTYIQSRFYRAPEVMMGARYGMPIDMWSLGCILAELLIGFPLLPGEDEADQMACIIELLGMPPPKLIEQGKRSKNFISSKGLPRYCTATTLADGTTVLSGGMSRRGKPRGPPGSKSFVTALKGCQDKFFIDFIRRCLEWDPEKRLTPAAALRHAWLRRRLPRAPHDDEPNHVSIAHHAKRLAHPNCTDK
Summary
Similarity
Belongs to the protein kinase superfamily.
Uniprot
A0A2H1V172
A0A3S2M7X3
A0A2W1B326
A0A194Q6M9
A0A222AJ63
A0A212F9D9
+ More
A0A1W4WP08 A0A1Y1L2W6 A0A0T6B1H5 A0A2P8XTB8 A0A2J7PXY8 A0A1B6CMA4 A0A0N7Z8L1 A0A023F4J6 A0A226E4U8 A0A1B6LKS8 E9IPV3 A0A3S2MCM3 A0A0A9X7G4 A0A0A9WZ53 A0A1B6HJ39 A0A3B4UQW6 A0A1A7WLF5 D6W776 A0A146M0X0 A0A1A8GV89 K1QR76 A0A146LZ47 A0A2R5LMB7 A0A1A8AR25 A0A226P041 A0A3Q4H6P9 A0A1A7Z2W2 A0A2C9JYG4 A0A1A8SBS8 A0A0S7H005 A0A3B4YFU7 A0A3B4USG0 A0A1A8RTA9 A0A226MAP3 A0A3Q3ILL2 A0A3Q4H6P4 A0A293LS30 A0A3P8PKD4 A0A1A8N1U1 A0A3P9K857 A0A3P9H8A0 I3KNL6 A0A1A8FXU7 H2MSF8 A0A3B3QG30 A0A0N0BFT8 A0A2T7NUD3 A0A076G429 A0A3M6T5X0 A0A3Q2GQ47 A0A2R7WCP2 A0A1A8UET4 A0A3B4G0D5 A0A3P8PK24 A0A3P9CVB4 A0A3Q2WDU6 A0A1A8CF19 A0A3P9CVB6 A0A226MP23 A0A2R8QCS7 V9KQC1 A0A3Q1JG40 A0A1A8I057 A0A131XUL6 Q5RHV3 W4ZFF0 A0A3B5B7V7 A0A3B3QIR8 A0A1S3JMR2 A0A0F8AS94 A0A3B3E0B5 A0A3Q3N826 F6T718 A0A0L8GT78 E0W1C0 A0A060Z9F1 A0A0R4ITS5 A0A3Q1C5H9 A0A2B4RNR8 A0A3B3QHX3 N6T9F7 A0A3M7RLJ2 A0A315VEH8 A0A3B5KC00 T1JLC7 A0A3Q1JSB3 A0A3Q2PIW0 D0VY19 W5NH23 W5NH21 A0A3B3XUW5 A0A3Q3G7H0
A0A1W4WP08 A0A1Y1L2W6 A0A0T6B1H5 A0A2P8XTB8 A0A2J7PXY8 A0A1B6CMA4 A0A0N7Z8L1 A0A023F4J6 A0A226E4U8 A0A1B6LKS8 E9IPV3 A0A3S2MCM3 A0A0A9X7G4 A0A0A9WZ53 A0A1B6HJ39 A0A3B4UQW6 A0A1A7WLF5 D6W776 A0A146M0X0 A0A1A8GV89 K1QR76 A0A146LZ47 A0A2R5LMB7 A0A1A8AR25 A0A226P041 A0A3Q4H6P9 A0A1A7Z2W2 A0A2C9JYG4 A0A1A8SBS8 A0A0S7H005 A0A3B4YFU7 A0A3B4USG0 A0A1A8RTA9 A0A226MAP3 A0A3Q3ILL2 A0A3Q4H6P4 A0A293LS30 A0A3P8PKD4 A0A1A8N1U1 A0A3P9K857 A0A3P9H8A0 I3KNL6 A0A1A8FXU7 H2MSF8 A0A3B3QG30 A0A0N0BFT8 A0A2T7NUD3 A0A076G429 A0A3M6T5X0 A0A3Q2GQ47 A0A2R7WCP2 A0A1A8UET4 A0A3B4G0D5 A0A3P8PK24 A0A3P9CVB4 A0A3Q2WDU6 A0A1A8CF19 A0A3P9CVB6 A0A226MP23 A0A2R8QCS7 V9KQC1 A0A3Q1JG40 A0A1A8I057 A0A131XUL6 Q5RHV3 W4ZFF0 A0A3B5B7V7 A0A3B3QIR8 A0A1S3JMR2 A0A0F8AS94 A0A3B3E0B5 A0A3Q3N826 F6T718 A0A0L8GT78 E0W1C0 A0A060Z9F1 A0A0R4ITS5 A0A3Q1C5H9 A0A2B4RNR8 A0A3B3QHX3 N6T9F7 A0A3M7RLJ2 A0A315VEH8 A0A3B5KC00 T1JLC7 A0A3Q1JSB3 A0A3Q2PIW0 D0VY19 W5NH23 W5NH21 A0A3B3XUW5 A0A3Q3G7H0
Pubmed
28756777
26354079
22118469
28004739
29403074
27129103
+ More
25474469 21282665 25401762 18362917 19820115 26823975 22992520 24621616 28717047 25186727 15562597 17554307 29240929 30382153 23594743 24402279 29652888 25835551 29451363 12481130 15114417 20566863 24755649 23537049 30375419 29703783 21551351
25474469 21282665 25401762 18362917 19820115 26823975 22992520 24621616 28717047 25186727 15562597 17554307 29240929 30382153 23594743 24402279 29652888 25835551 29451363 12481130 15114417 20566863 24755649 23537049 30375419 29703783 21551351
EMBL
ODYU01000028
SOQ34102.1
RSAL01000012
RVE53496.1
KZ150400
PZC71019.1
+ More
KQ459460 KPJ00660.1 KY609556 ASO76420.1 AGBW02009612 OWR50329.1 GEZM01066361 JAV67924.1 LJIG01016290 KRT81044.1 PYGN01001379 PSN35261.1 NEVH01020852 PNF21201.1 GEDC01022783 JAS14515.1 GDKW01003043 JAI53552.1 GBBI01002550 JAC16162.1 LNIX01000006 OXA52745.1 GEBQ01015687 JAT24290.1 GL764626 EFZ17408.1 CM012459 RVE56489.1 GBHO01030579 GBHO01030578 JAG13025.1 JAG13026.1 GBHO01030580 JAG13024.1 GECU01033019 JAS74687.1 HADW01005155 SBP06555.1 KQ971307 EFA11400.2 GDHC01006103 JAQ12526.1 HAEC01006806 SBQ74944.1 JH818808 EKC39457.1 GDHC01006723 JAQ11906.1 GGLE01006544 MBY10670.1 HADY01018195 SBP56680.1 AWGT02000238 AWGT02000137 OXB72972.1 OXB75741.1 HADX01014288 SBP36520.1 HAEI01012929 SBS15398.1 GBYX01447246 JAO34200.1 HAEH01019254 SBS08947.1 AWGT02012092 OXB52278.1 GFWV01004910 MAA29640.1 HAEF01021889 SBR63048.1 AERX01019352 AERX01019353 AERX01019354 AERX01019355 AERX01019356 HAEB01017166 SBQ63693.1 KQ435794 KOX73804.1 PZQS01000009 PVD24771.1 KJ784521 AII25879.1 RCHS01004263 RMX36674.1 KK854551 PTY16790.1 HAEJ01005345 SBS45802.1 HADZ01013409 SBP77350.1 MCFN01000578 OXB57076.1 BX465210 JW868026 AFP00544.1 HAED01004171 SBQ90201.1 GEFM01006450 GEGO01001631 JAP69346.1 JAR93773.1 BC139631 AAI39632.1 AAGJ04006022 KQ040995 KKF32165.1 KQ420477 KOF80153.1 DS235867 EEB19426.1 FR936617 CDQ98319.1 LSMT01000442 PFX17915.1 APGK01039173 KB740967 KB632399 ENN76889.1 ERL94622.1 REGN01003122 RNA24416.1 NHOQ01001892 PWA21811.1 JH430326 AB490373 BAI50371.1 AHAT01008866
KQ459460 KPJ00660.1 KY609556 ASO76420.1 AGBW02009612 OWR50329.1 GEZM01066361 JAV67924.1 LJIG01016290 KRT81044.1 PYGN01001379 PSN35261.1 NEVH01020852 PNF21201.1 GEDC01022783 JAS14515.1 GDKW01003043 JAI53552.1 GBBI01002550 JAC16162.1 LNIX01000006 OXA52745.1 GEBQ01015687 JAT24290.1 GL764626 EFZ17408.1 CM012459 RVE56489.1 GBHO01030579 GBHO01030578 JAG13025.1 JAG13026.1 GBHO01030580 JAG13024.1 GECU01033019 JAS74687.1 HADW01005155 SBP06555.1 KQ971307 EFA11400.2 GDHC01006103 JAQ12526.1 HAEC01006806 SBQ74944.1 JH818808 EKC39457.1 GDHC01006723 JAQ11906.1 GGLE01006544 MBY10670.1 HADY01018195 SBP56680.1 AWGT02000238 AWGT02000137 OXB72972.1 OXB75741.1 HADX01014288 SBP36520.1 HAEI01012929 SBS15398.1 GBYX01447246 JAO34200.1 HAEH01019254 SBS08947.1 AWGT02012092 OXB52278.1 GFWV01004910 MAA29640.1 HAEF01021889 SBR63048.1 AERX01019352 AERX01019353 AERX01019354 AERX01019355 AERX01019356 HAEB01017166 SBQ63693.1 KQ435794 KOX73804.1 PZQS01000009 PVD24771.1 KJ784521 AII25879.1 RCHS01004263 RMX36674.1 KK854551 PTY16790.1 HAEJ01005345 SBS45802.1 HADZ01013409 SBP77350.1 MCFN01000578 OXB57076.1 BX465210 JW868026 AFP00544.1 HAED01004171 SBQ90201.1 GEFM01006450 GEGO01001631 JAP69346.1 JAR93773.1 BC139631 AAI39632.1 AAGJ04006022 KQ040995 KKF32165.1 KQ420477 KOF80153.1 DS235867 EEB19426.1 FR936617 CDQ98319.1 LSMT01000442 PFX17915.1 APGK01039173 KB740967 KB632399 ENN76889.1 ERL94622.1 REGN01003122 RNA24416.1 NHOQ01001892 PWA21811.1 JH430326 AB490373 BAI50371.1 AHAT01008866
Proteomes
UP000283053
UP000053268
UP000007151
UP000192223
UP000245037
UP000235965
+ More
UP000198287 UP000261420 UP000007266 UP000005408 UP000198419 UP000261580 UP000076420 UP000261360 UP000261600 UP000265100 UP000265180 UP000265200 UP000005207 UP000001038 UP000261540 UP000053105 UP000245119 UP000275408 UP000265020 UP000261460 UP000265160 UP000264840 UP000198323 UP000000437 UP000265040 UP000007110 UP000261400 UP000085678 UP000261560 UP000261640 UP000008144 UP000053454 UP000009046 UP000193380 UP000257160 UP000225706 UP000019118 UP000030742 UP000276133 UP000005226 UP000265000 UP000018468 UP000261480 UP000261660
UP000198287 UP000261420 UP000007266 UP000005408 UP000198419 UP000261580 UP000076420 UP000261360 UP000261600 UP000265100 UP000265180 UP000265200 UP000005207 UP000001038 UP000261540 UP000053105 UP000245119 UP000275408 UP000265020 UP000261460 UP000265160 UP000264840 UP000198323 UP000000437 UP000265040 UP000007110 UP000261400 UP000085678 UP000261560 UP000261640 UP000008144 UP000053454 UP000009046 UP000193380 UP000257160 UP000225706 UP000019118 UP000030742 UP000276133 UP000005226 UP000265000 UP000018468 UP000261480 UP000261660
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
A0A2H1V172
A0A3S2M7X3
A0A2W1B326
A0A194Q6M9
A0A222AJ63
A0A212F9D9
+ More
A0A1W4WP08 A0A1Y1L2W6 A0A0T6B1H5 A0A2P8XTB8 A0A2J7PXY8 A0A1B6CMA4 A0A0N7Z8L1 A0A023F4J6 A0A226E4U8 A0A1B6LKS8 E9IPV3 A0A3S2MCM3 A0A0A9X7G4 A0A0A9WZ53 A0A1B6HJ39 A0A3B4UQW6 A0A1A7WLF5 D6W776 A0A146M0X0 A0A1A8GV89 K1QR76 A0A146LZ47 A0A2R5LMB7 A0A1A8AR25 A0A226P041 A0A3Q4H6P9 A0A1A7Z2W2 A0A2C9JYG4 A0A1A8SBS8 A0A0S7H005 A0A3B4YFU7 A0A3B4USG0 A0A1A8RTA9 A0A226MAP3 A0A3Q3ILL2 A0A3Q4H6P4 A0A293LS30 A0A3P8PKD4 A0A1A8N1U1 A0A3P9K857 A0A3P9H8A0 I3KNL6 A0A1A8FXU7 H2MSF8 A0A3B3QG30 A0A0N0BFT8 A0A2T7NUD3 A0A076G429 A0A3M6T5X0 A0A3Q2GQ47 A0A2R7WCP2 A0A1A8UET4 A0A3B4G0D5 A0A3P8PK24 A0A3P9CVB4 A0A3Q2WDU6 A0A1A8CF19 A0A3P9CVB6 A0A226MP23 A0A2R8QCS7 V9KQC1 A0A3Q1JG40 A0A1A8I057 A0A131XUL6 Q5RHV3 W4ZFF0 A0A3B5B7V7 A0A3B3QIR8 A0A1S3JMR2 A0A0F8AS94 A0A3B3E0B5 A0A3Q3N826 F6T718 A0A0L8GT78 E0W1C0 A0A060Z9F1 A0A0R4ITS5 A0A3Q1C5H9 A0A2B4RNR8 A0A3B3QHX3 N6T9F7 A0A3M7RLJ2 A0A315VEH8 A0A3B5KC00 T1JLC7 A0A3Q1JSB3 A0A3Q2PIW0 D0VY19 W5NH23 W5NH21 A0A3B3XUW5 A0A3Q3G7H0
A0A1W4WP08 A0A1Y1L2W6 A0A0T6B1H5 A0A2P8XTB8 A0A2J7PXY8 A0A1B6CMA4 A0A0N7Z8L1 A0A023F4J6 A0A226E4U8 A0A1B6LKS8 E9IPV3 A0A3S2MCM3 A0A0A9X7G4 A0A0A9WZ53 A0A1B6HJ39 A0A3B4UQW6 A0A1A7WLF5 D6W776 A0A146M0X0 A0A1A8GV89 K1QR76 A0A146LZ47 A0A2R5LMB7 A0A1A8AR25 A0A226P041 A0A3Q4H6P9 A0A1A7Z2W2 A0A2C9JYG4 A0A1A8SBS8 A0A0S7H005 A0A3B4YFU7 A0A3B4USG0 A0A1A8RTA9 A0A226MAP3 A0A3Q3ILL2 A0A3Q4H6P4 A0A293LS30 A0A3P8PKD4 A0A1A8N1U1 A0A3P9K857 A0A3P9H8A0 I3KNL6 A0A1A8FXU7 H2MSF8 A0A3B3QG30 A0A0N0BFT8 A0A2T7NUD3 A0A076G429 A0A3M6T5X0 A0A3Q2GQ47 A0A2R7WCP2 A0A1A8UET4 A0A3B4G0D5 A0A3P8PK24 A0A3P9CVB4 A0A3Q2WDU6 A0A1A8CF19 A0A3P9CVB6 A0A226MP23 A0A2R8QCS7 V9KQC1 A0A3Q1JG40 A0A1A8I057 A0A131XUL6 Q5RHV3 W4ZFF0 A0A3B5B7V7 A0A3B3QIR8 A0A1S3JMR2 A0A0F8AS94 A0A3B3E0B5 A0A3Q3N826 F6T718 A0A0L8GT78 E0W1C0 A0A060Z9F1 A0A0R4ITS5 A0A3Q1C5H9 A0A2B4RNR8 A0A3B3QHX3 N6T9F7 A0A3M7RLJ2 A0A315VEH8 A0A3B5KC00 T1JLC7 A0A3Q1JSB3 A0A3Q2PIW0 D0VY19 W5NH23 W5NH21 A0A3B3XUW5 A0A3Q3G7H0
PDB
4AZF
E-value=1.62383e-74,
Score=708
Ontologies
GO
Topology
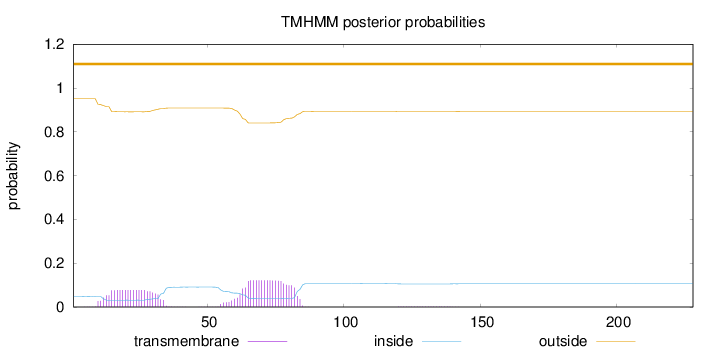
Length:
228
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.23594000000001
Exp number, first 60 AAs:
1.7651
Total prob of N-in:
0.04893
outside
1 - 228
Population Genetic Test Statistics
Pi
217.643922
Theta
168.11098
Tajima's D
0.230012
CLR
0.825936
CSRT
0.435778211089446
Interpretation
Uncertain
