Gene
KWMTBOMO15992
Pre Gene Modal
BGIBMGA005726
Annotation
PREDICTED:_probable_cation-transporting_ATPase_13A3_[Bombyx_mori]
Full name
Cation-transporting ATPase
Location in the cell
PlasmaMembrane Reliability : 4.437
Sequence
CDS
ATGACCTGTGTGAGATGCAGTTTCCAGGTTGCCGATCAGGTTTTAATTGTTGAAACATATCAAAAGAAATGTAAAAGTTATTATATTCACAAAATACGCCATAAAAACATAGAAGACACAAGTGGATTATTCAATGAGGGTTTTGATGTTGAGCCGGGAGATGATAAAAGAAACTTTGAAACTGCTGATGCTATACACACTCCCATACATTTGGACGATGGTTCCACTCTTAATGTCCAAAACTACCGTTATTTCCGGCACAAAAAGCAGTCTATAATATGGGACGCGGGTAGAGCGCAATGGGTGAGACTAGCTGGAATCGAAGCTGGGGCGACATGTGCGCAACTTCATGCAAGGGGCGCCATAGCACCTACTAGTGAAAAAAGGATTAGGATGTTAAATATATACGGCTTGAACGAGATAAAGGTTCCTGTACAGTCTATCTTCACTTTGATCCTCTTGGAAGTATTCAATCCGTTCTACGTATTCCAATTTTTCACAATAGCCGTGTGGCTCGCAGAACCTTACTATTATTATTGTATTGCCGTTGTTTTAATGTCGACGTTCGGTGTTGCCACTTCTGTGATACAAACCAAAAAGAACCAGGAGAGTCTTCGCGCGACCGTGGAGGCTCACAGCACGGTGCGGCTGTGGGACGGGCGGGTGGTGGCGACGCGGGCAGTTTGTCCGGGGGACGTGGTACTGCTCCCGGCAAGGGGCTGCACGCTGCATTTCGACGCGCTGCTGCTCACCGGCACCGCCGTGCTCAACGAGAGCATGCTCACCGGTGAATCAGTGCCGGTCGGGAAAACGGCTTTGTTGAGGGTCGACAAACCATTCGACATGAAGGAGCACGCGGCCAGCATTCTGTTCTGCGGCACCAGAGTCATTCAGACCCGGTACTACAACAACGAACCTGTCAGGGCCTTGGTGCTGCGTACGGGCTACAACACAAGTAAAGGGCAGCTGGTCCGCAGTATACTGTACCCGGTGCCGGCGGACTTCAAGTTCGACCGGGACTCGTACAAGTTCATCATCATCCTAGCCGGGATAGCGGCGTGCGGACTGGCCTATACCGTGGCCTTGAAGGCGTACCGCGCTCTCACTCCCAGCGACATAGTCCTGAAGGCGCTGGACATCATCACGATCGTGATCCCGCCCGCGCTGCCGGCGGCCATGACGGTGGGGCGGCTGTACGCCGTGGCGCGGCTGGCGAGCGCGCGCGTCGCCTGCCTCAACACGCGCGCCGTCAACGTCACCGGCTCGCTCGACTGCATGTGCTTCGACAAGACAGGGACGTTAACAGAGGACGGGCTAGACATGTGGGGCGTGGTGCCGGTGAGCGCGGCCACCGACCCGCCGATGCTGGGCCGCCCGCAGCGGGACCCCGCCCTCATCAACGACTTGCACGACCTCAAGATGGGCATGGCCGTCTGCCACAGCCTCACGCTGCTCGACGGGGAGCTCGCCGGAGACCCGCTGGACTTGAAGATGTTCGAGTCGACGGGCTGGCTGCTGGAGGAGCCCGACGTGCCCGAGCCGTCCAACTACGAGGTTCTGACTCCGACCATAGTGAGACCCAAGAAAACTGCAAACATCAACGTCGATGACATTCATATGCCGCTGGAGCTGGGCATAGTGCAGCAGTATCAGTTCGTCTCGTCACTCCAGAGGTCCTCGGTCGTGGTCCGGATGCTGGGAGAAGATGTACTCCGCGTTTACTGTAAAGGAGCTCCCGAGATGATTACGACGCTGTGCCGACTCGAGACAGTTCCAAATAATCTAAGCGACGTTCTCGCCTCGTATGCGGAGAAGGGTTACCGTGTCATCGCGATGTCTACCCGGATAATGGAAGCGACGCTCAAGCAGATTCACAAGATGACAAGAGAAGAGGCCGAACGTGATCTAGAATTCTTAGGGCTCGTCATAATGGAGAATCGTCTGAAAGCTGCGACCACGGGCATCATACGAGAACTCAAGGAAGCCAATATACATGTGGTCATGATAACAGGGGACAACGTGCACACGGCGGTGAGCGTCGCCAAGGAGTGCGGCATCCTCGCCAGCGGGGAGCGCATCGTGCAGCTGAGCGGTGACGCCGCCGCCCTGTACTGCGAGAGCACCCTGCACGGGGCGCCGGTGGGGCGACGTACGTTGCTCCAGCGCGAATGGCGCAGTGAGGACAGCGGGCGCGGCTCCTGGGGACAGTCCGGCTCCGCTTCAGGCTCGGGCTTTGCCCCCAACGACCCTGAGGCTCCGGCCTCCGAACCTCGCTACAAGATTGTCGTAACTGGTGATACCTGGAGGATCCTGCGCGAGCACTACCCCGACATGGTGGGGCGCGTCGTGGAGCGCGGCGCCGTGTTCGCGAGGATGTCGTCCGATCAGAAGCAGCAGCTCATCGTTGAATATCAGGCTCTGGGATATTACGTCGGCATGTGCGGCGACGGCGCCAACGACTGCGGCGCCCTGCGCGCGGCGCACTGCGGCGTGTCGCTGTCGGAGGCGGAGAGCAGCGTGGCGGCCAGCTTCACGGCGCGCGAGCTGTCCGCCGCGCCGCGCCTGCTGCGGGAGGGCCGCGCCGCGCTCGCCACCAGCTTCGGACTCTTCAAGTTCATGGTGGGCTACTCGCTCACGGAGTTCTTCTCCGTCGCCTTCCTCTACTACTTCGACTCGAACCTGACCGACTTCCAGTTCCTGTACATCGACATCGCGTTGATCGTCAATTTCGCCTTCTTCTTCGGCATGACTGAGGCCCACTCGGGCAAGTTGTGCGAGAGGCCCCCGCTGACGTCCCTGCTGGGCCTGGTGCCGCTGTGCTCGCTGGTGGGACAGCTGTCGTTAATCGCGGTGGCGCAGTACTTCGGCTACTACGCCCTGACGTTCTTCCCGTGGTACGTGAGGCATACGTACGCCGGCGAGGAAGAAAACGAGTGCTGGGAGAACTACGTGATCTTCGCGATATCGATGTTCCAGTACGTGATAATGGCGATCGTGTTCAGCCACGGCGCGCCGCACCGCCGCTCGATCCTCAGCAACAGAAAGTTCGTGGCGAGTCTGCTGGTGATGACGTCACTGTGCGTGTACATAGCCGTGTCGCCGGCCGCCTGGGTGGCTTCGTTCCTGCAGCTCCGGATGCCGAAGGACGTCCTGCTGTGCTACCTGGTGCTGGCCATCACGGCAGCCAACTTCGTCGTGGCGCTGCTGTTCGAAAGATTGGTCATAGAGTACCTAATGGAAGGCAGAGAGATGATTCCGAAATTTTTGAAGGAAAAACGAGTTAGGAAGACTCCTCACTTGATGCTGAAAAGGCAGCTGACTGACATGGACTACCTGGATTGCGACAGCCTCGTGAAGTTTGATAAGGAGGGTGCCATTTCGGATGCCAGTTCCGGCAGAGGTAGCTGA
Protein
MTCVRCSFQVADQVLIVETYQKKCKSYYIHKIRHKNIEDTSGLFNEGFDVEPGDDKRNFETADAIHTPIHLDDGSTLNVQNYRYFRHKKQSIIWDAGRAQWVRLAGIEAGATCAQLHARGAIAPTSEKRIRMLNIYGLNEIKVPVQSIFTLILLEVFNPFYVFQFFTIAVWLAEPYYYYCIAVVLMSTFGVATSVIQTKKNQESLRATVEAHSTVRLWDGRVVATRAVCPGDVVLLPARGCTLHFDALLLTGTAVLNESMLTGESVPVGKTALLRVDKPFDMKEHAASILFCGTRVIQTRYYNNEPVRALVLRTGYNTSKGQLVRSILYPVPADFKFDRDSYKFIIILAGIAACGLAYTVALKAYRALTPSDIVLKALDIITIVIPPALPAAMTVGRLYAVARLASARVACLNTRAVNVTGSLDCMCFDKTGTLTEDGLDMWGVVPVSAATDPPMLGRPQRDPALINDLHDLKMGMAVCHSLTLLDGELAGDPLDLKMFESTGWLLEEPDVPEPSNYEVLTPTIVRPKKTANINVDDIHMPLELGIVQQYQFVSSLQRSSVVVRMLGEDVLRVYCKGAPEMITTLCRLETVPNNLSDVLASYAEKGYRVIAMSTRIMEATLKQIHKMTREEAERDLEFLGLVIMENRLKAATTGIIRELKEANIHVVMITGDNVHTAVSVAKECGILASGERIVQLSGDAAALYCESTLHGAPVGRRTLLQREWRSEDSGRGSWGQSGSASGSGFAPNDPEAPASEPRYKIVVTGDTWRILREHYPDMVGRVVERGAVFARMSSDQKQQLIVEYQALGYYVGMCGDGANDCGALRAAHCGVSLSEAESSVAASFTARELSAAPRLLREGRAALATSFGLFKFMVGYSLTEFFSVAFLYYFDSNLTDFQFLYIDIALIVNFAFFFGMTEAHSGKLCERPPLTSLLGLVPLCSLVGQLSLIAVAQYFGYYALTFFPWYVRHTYAGEEENECWENYVIFAISMFQYVIMAIVFSHGAPHRRSILSNRKFVASLLVMTSLCVYIAVSPAAWVASFLQLRMPKDVLLCYLVLAITAANFVVALLFERLVIEYLMEGREMIPKFLKEKRVRKTPHLMLKRQLTDMDYLDCDSLVKFDKEGAISDASSGRGS
Summary
Similarity
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type V subfamily.
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family.
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family.
Uniprot
H9J884
A0A3S2LGR5
A0A2H1VY27
A0A212FLP4
A0A2H1VQ27
A0A0L7L9C2
+ More
A0A0L7RF13 A0A1B6J8B3 A0A0C9QI04 E2A6U0 A0A1B6H2H5 A0A154PIT9 A0A0C9QR79 A0A2A3EBA6 V9I9W4 V9IAS5 A0A1B6M5K1 A0A088ARC9 A0A067RJ75 A0A2J7RFI4 A0A067QEM5 A0A2H8TIA6 E9J5D4 A0A139WGX1 A0A151I0S0 A0A2S2NEI9 A0A2S2Q7K1 X1WP34 A0A195EYL9 A0A0M8ZZJ9 K7JAM2 A0A069DXS7 A0A195C3K6 A0A151X5X3 A0A151J619 T1HUM0 A0A2P8YLJ0 A0A3L8D579 A0A195EYI3 A0A151IA01 E0VGV9 A0A154P353 A0A2H8TTR1 J9KAS7 A0A2S2P820 T1HPU5 A0A023F2U2 A0A224XGH5 A0A069DYB5 T1K1X3 Q16XE5 A0A1S4FKQ2 A0A2R5L8D8 A0A0A9XGH2 A0A146LYI6 A0A1Y1K832 A0A146M0L3 A0A1Q3F0M6 A0A0N8CR08 A0A0P5SGE5 A0A0P6BE48 A0A0N7ZJH2 A0A0P5BQB4 A0A1V9XBK9 A0A0N7ZTS8 A0A162PFK4 A0A293N079 A0A0P5SVJ5 A0A026W8N8 A0A0K0EYM5 A0A0P5CWW2 A0A0N5C4K8 A0A0P4Z1I1 A0A0K0E871 F1KSX7 F1KSF7 A0A090L2Y3 A0A3B3RF51 A0A3B3RHI3 A0A3B3RFR1 A0A3B3REZ2 A0A3B3RF49 M3ZTQ6 A0A158PSU0 A0A0H5SLJ0 A0A3P7DE29 A0A0K0JMA7 A0A3Q2Y913 A0A3Q2Y7L2 A0A3B5PRK5 A0A1D1W8N8 A0A0R3QL57 A0A3B5PQT5 A0A3P6SR81
A0A0L7RF13 A0A1B6J8B3 A0A0C9QI04 E2A6U0 A0A1B6H2H5 A0A154PIT9 A0A0C9QR79 A0A2A3EBA6 V9I9W4 V9IAS5 A0A1B6M5K1 A0A088ARC9 A0A067RJ75 A0A2J7RFI4 A0A067QEM5 A0A2H8TIA6 E9J5D4 A0A139WGX1 A0A151I0S0 A0A2S2NEI9 A0A2S2Q7K1 X1WP34 A0A195EYL9 A0A0M8ZZJ9 K7JAM2 A0A069DXS7 A0A195C3K6 A0A151X5X3 A0A151J619 T1HUM0 A0A2P8YLJ0 A0A3L8D579 A0A195EYI3 A0A151IA01 E0VGV9 A0A154P353 A0A2H8TTR1 J9KAS7 A0A2S2P820 T1HPU5 A0A023F2U2 A0A224XGH5 A0A069DYB5 T1K1X3 Q16XE5 A0A1S4FKQ2 A0A2R5L8D8 A0A0A9XGH2 A0A146LYI6 A0A1Y1K832 A0A146M0L3 A0A1Q3F0M6 A0A0N8CR08 A0A0P5SGE5 A0A0P6BE48 A0A0N7ZJH2 A0A0P5BQB4 A0A1V9XBK9 A0A0N7ZTS8 A0A162PFK4 A0A293N079 A0A0P5SVJ5 A0A026W8N8 A0A0K0EYM5 A0A0P5CWW2 A0A0N5C4K8 A0A0P4Z1I1 A0A0K0E871 F1KSX7 F1KSF7 A0A090L2Y3 A0A3B3RF51 A0A3B3RHI3 A0A3B3RFR1 A0A3B3REZ2 A0A3B3RF49 M3ZTQ6 A0A158PSU0 A0A0H5SLJ0 A0A3P7DE29 A0A0K0JMA7 A0A3Q2Y913 A0A3Q2Y7L2 A0A3B5PRK5 A0A1D1W8N8 A0A0R3QL57 A0A3B5PQT5 A0A3P6SR81
EC Number
3.6.3.-
Pubmed
EMBL
BABH01038514
BABH01038515
BABH01038516
RSAL01000012
RVE53498.1
ODYU01005156
+ More
SOQ45741.1 AGBW02007724 OWR54665.1 ODYU01003767 SOQ42945.1 JTDY01002168 KOB71964.1 KQ414608 KOC69420.1 GECU01012353 JAS95353.1 GBYB01003169 JAG72936.1 GL437203 EFN70806.1 GECZ01022071 GECZ01000908 JAS47698.1 JAS68861.1 KQ434931 KZC11791.1 GBYB01003167 JAG72934.1 KZ288308 PBC28744.1 JR036862 JR036864 AEY57435.1 AEY57437.1 JR036861 JR036863 AEY57434.1 AEY57436.1 GEBQ01031444 GEBQ01027597 GEBQ01008777 JAT08533.1 JAT12380.1 JAT31200.1 KK852439 KDR23872.1 NEVH01004413 PNF39586.1 KK853768 KDQ84122.1 GFXV01001946 MBW13751.1 GL768143 EFZ11970.1 KQ971344 KYB27139.1 KQ976609 KYM79315.1 GGMR01002936 MBY15555.1 GGMS01003969 MBY73172.1 ABLF02014735 ABLF02014736 KQ981905 KYN33318.1 KQ435791 KOX74320.1 AAZX01017245 GBGD01000199 JAC88690.1 KQ978344 KYM94771.1 KQ982494 KYQ55744.1 KQ979928 KYN18606.1 ACPB03005214 PYGN01000511 PSN45084.1 QOIP01000013 RLU15384.1 KQ981923 KYN32967.1 KQ978249 KYM96005.1 AAZO01002251 DS235152 EEB12615.1 KQ434796 KZC05774.1 GFXV01005811 MBW17616.1 ABLF02038923 ABLF02038926 ABLF02038932 GGMR01012951 MBY25570.1 ACPB03008263 GBBI01003199 JAC15513.1 GFTR01008734 JAW07692.1 GBGD01000117 JAC88772.1 CAEY01001357 CH477540 EAT39291.1 GGLE01001640 MBY05766.1 GBHO01025693 GBHO01025692 GBHO01025691 GBHO01017614 GBRD01009067 JAG17911.1 JAG17912.1 JAG17913.1 JAG25990.1 JAG56754.1 GDHC01005978 JAQ12651.1 GEZM01089272 JAV57563.1 GDHC01006233 JAQ12396.1 GFDL01013932 JAV21113.1 GDIP01107383 JAL96331.1 GDIP01140381 JAL63333.1 GDIP01030692 JAM73023.1 GDIP01239361 JAI84040.1 GDIP01185372 JAJ38030.1 MNPL01015760 OQR70924.1 GDIP01213313 JAJ10089.1 LRGB01000512 KZS18804.1 GFWV01021745 MAA46473.1 GDIP01134670 JAL69044.1 KK107335 EZA52462.1 GDIP01164314 JAJ59088.1 GDIP01222062 JAJ01340.1 JI165340 ADY40981.1 JI165114 ADY40811.1 LN609528 CEF64067.1 UZAD01013524 VDN95306.1 LN856968 CRZ24617.1 UYWW01000295 VDM08030.1 BDGG01000018 GAV08668.1 UZAG01015606 VDO21868.1 UYRX01000209 VDK77556.1
SOQ45741.1 AGBW02007724 OWR54665.1 ODYU01003767 SOQ42945.1 JTDY01002168 KOB71964.1 KQ414608 KOC69420.1 GECU01012353 JAS95353.1 GBYB01003169 JAG72936.1 GL437203 EFN70806.1 GECZ01022071 GECZ01000908 JAS47698.1 JAS68861.1 KQ434931 KZC11791.1 GBYB01003167 JAG72934.1 KZ288308 PBC28744.1 JR036862 JR036864 AEY57435.1 AEY57437.1 JR036861 JR036863 AEY57434.1 AEY57436.1 GEBQ01031444 GEBQ01027597 GEBQ01008777 JAT08533.1 JAT12380.1 JAT31200.1 KK852439 KDR23872.1 NEVH01004413 PNF39586.1 KK853768 KDQ84122.1 GFXV01001946 MBW13751.1 GL768143 EFZ11970.1 KQ971344 KYB27139.1 KQ976609 KYM79315.1 GGMR01002936 MBY15555.1 GGMS01003969 MBY73172.1 ABLF02014735 ABLF02014736 KQ981905 KYN33318.1 KQ435791 KOX74320.1 AAZX01017245 GBGD01000199 JAC88690.1 KQ978344 KYM94771.1 KQ982494 KYQ55744.1 KQ979928 KYN18606.1 ACPB03005214 PYGN01000511 PSN45084.1 QOIP01000013 RLU15384.1 KQ981923 KYN32967.1 KQ978249 KYM96005.1 AAZO01002251 DS235152 EEB12615.1 KQ434796 KZC05774.1 GFXV01005811 MBW17616.1 ABLF02038923 ABLF02038926 ABLF02038932 GGMR01012951 MBY25570.1 ACPB03008263 GBBI01003199 JAC15513.1 GFTR01008734 JAW07692.1 GBGD01000117 JAC88772.1 CAEY01001357 CH477540 EAT39291.1 GGLE01001640 MBY05766.1 GBHO01025693 GBHO01025692 GBHO01025691 GBHO01017614 GBRD01009067 JAG17911.1 JAG17912.1 JAG17913.1 JAG25990.1 JAG56754.1 GDHC01005978 JAQ12651.1 GEZM01089272 JAV57563.1 GDHC01006233 JAQ12396.1 GFDL01013932 JAV21113.1 GDIP01107383 JAL96331.1 GDIP01140381 JAL63333.1 GDIP01030692 JAM73023.1 GDIP01239361 JAI84040.1 GDIP01185372 JAJ38030.1 MNPL01015760 OQR70924.1 GDIP01213313 JAJ10089.1 LRGB01000512 KZS18804.1 GFWV01021745 MAA46473.1 GDIP01134670 JAL69044.1 KK107335 EZA52462.1 GDIP01164314 JAJ59088.1 GDIP01222062 JAJ01340.1 JI165340 ADY40981.1 JI165114 ADY40811.1 LN609528 CEF64067.1 UZAD01013524 VDN95306.1 LN856968 CRZ24617.1 UYWW01000295 VDM08030.1 BDGG01000018 GAV08668.1 UZAG01015606 VDO21868.1 UYRX01000209 VDK77556.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000037510
UP000053825
UP000000311
+ More
UP000076502 UP000242457 UP000005203 UP000027135 UP000235965 UP000007266 UP000078540 UP000007819 UP000078541 UP000053105 UP000002358 UP000078542 UP000075809 UP000078492 UP000015103 UP000245037 UP000279307 UP000009046 UP000015104 UP000008820 UP000192247 UP000076858 UP000053097 UP000035680 UP000046392 UP000035681 UP000035682 UP000261540 UP000002852 UP000038020 UP000278627 UP000270924 UP000006672 UP000264820 UP000186922 UP000050602 UP000277928
UP000076502 UP000242457 UP000005203 UP000027135 UP000235965 UP000007266 UP000078540 UP000007819 UP000078541 UP000053105 UP000002358 UP000078542 UP000075809 UP000078492 UP000015103 UP000245037 UP000279307 UP000009046 UP000015104 UP000008820 UP000192247 UP000076858 UP000053097 UP000035680 UP000046392 UP000035681 UP000035682 UP000261540 UP000002852 UP000038020 UP000278627 UP000270924 UP000006672 UP000264820 UP000186922 UP000050602 UP000277928
PRIDE
Interpro
IPR036412
HAD-like_sf
+ More
IPR023299 ATPase_P-typ_cyto_dom_N
IPR006544 P-type_TPase_V
IPR023298 ATPase_P-typ_TM_dom_sf
IPR001757 P_typ_ATPase
IPR018303 ATPase_P-typ_P_site
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR023214 HAD_sf
IPR004014 ATPase_P-typ_cation-transptr_N
IPR000961 AGC-kinase_C
IPR006068 ATPase_P-typ_cation-transptr_C
IPR023299 ATPase_P-typ_cyto_dom_N
IPR006544 P-type_TPase_V
IPR023298 ATPase_P-typ_TM_dom_sf
IPR001757 P_typ_ATPase
IPR018303 ATPase_P-typ_P_site
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR023214 HAD_sf
IPR004014 ATPase_P-typ_cation-transptr_N
IPR000961 AGC-kinase_C
IPR006068 ATPase_P-typ_cation-transptr_C
Gene 3D
ProteinModelPortal
H9J884
A0A3S2LGR5
A0A2H1VY27
A0A212FLP4
A0A2H1VQ27
A0A0L7L9C2
+ More
A0A0L7RF13 A0A1B6J8B3 A0A0C9QI04 E2A6U0 A0A1B6H2H5 A0A154PIT9 A0A0C9QR79 A0A2A3EBA6 V9I9W4 V9IAS5 A0A1B6M5K1 A0A088ARC9 A0A067RJ75 A0A2J7RFI4 A0A067QEM5 A0A2H8TIA6 E9J5D4 A0A139WGX1 A0A151I0S0 A0A2S2NEI9 A0A2S2Q7K1 X1WP34 A0A195EYL9 A0A0M8ZZJ9 K7JAM2 A0A069DXS7 A0A195C3K6 A0A151X5X3 A0A151J619 T1HUM0 A0A2P8YLJ0 A0A3L8D579 A0A195EYI3 A0A151IA01 E0VGV9 A0A154P353 A0A2H8TTR1 J9KAS7 A0A2S2P820 T1HPU5 A0A023F2U2 A0A224XGH5 A0A069DYB5 T1K1X3 Q16XE5 A0A1S4FKQ2 A0A2R5L8D8 A0A0A9XGH2 A0A146LYI6 A0A1Y1K832 A0A146M0L3 A0A1Q3F0M6 A0A0N8CR08 A0A0P5SGE5 A0A0P6BE48 A0A0N7ZJH2 A0A0P5BQB4 A0A1V9XBK9 A0A0N7ZTS8 A0A162PFK4 A0A293N079 A0A0P5SVJ5 A0A026W8N8 A0A0K0EYM5 A0A0P5CWW2 A0A0N5C4K8 A0A0P4Z1I1 A0A0K0E871 F1KSX7 F1KSF7 A0A090L2Y3 A0A3B3RF51 A0A3B3RHI3 A0A3B3RFR1 A0A3B3REZ2 A0A3B3RF49 M3ZTQ6 A0A158PSU0 A0A0H5SLJ0 A0A3P7DE29 A0A0K0JMA7 A0A3Q2Y913 A0A3Q2Y7L2 A0A3B5PRK5 A0A1D1W8N8 A0A0R3QL57 A0A3B5PQT5 A0A3P6SR81
A0A0L7RF13 A0A1B6J8B3 A0A0C9QI04 E2A6U0 A0A1B6H2H5 A0A154PIT9 A0A0C9QR79 A0A2A3EBA6 V9I9W4 V9IAS5 A0A1B6M5K1 A0A088ARC9 A0A067RJ75 A0A2J7RFI4 A0A067QEM5 A0A2H8TIA6 E9J5D4 A0A139WGX1 A0A151I0S0 A0A2S2NEI9 A0A2S2Q7K1 X1WP34 A0A195EYL9 A0A0M8ZZJ9 K7JAM2 A0A069DXS7 A0A195C3K6 A0A151X5X3 A0A151J619 T1HUM0 A0A2P8YLJ0 A0A3L8D579 A0A195EYI3 A0A151IA01 E0VGV9 A0A154P353 A0A2H8TTR1 J9KAS7 A0A2S2P820 T1HPU5 A0A023F2U2 A0A224XGH5 A0A069DYB5 T1K1X3 Q16XE5 A0A1S4FKQ2 A0A2R5L8D8 A0A0A9XGH2 A0A146LYI6 A0A1Y1K832 A0A146M0L3 A0A1Q3F0M6 A0A0N8CR08 A0A0P5SGE5 A0A0P6BE48 A0A0N7ZJH2 A0A0P5BQB4 A0A1V9XBK9 A0A0N7ZTS8 A0A162PFK4 A0A293N079 A0A0P5SVJ5 A0A026W8N8 A0A0K0EYM5 A0A0P5CWW2 A0A0N5C4K8 A0A0P4Z1I1 A0A0K0E871 F1KSX7 F1KSF7 A0A090L2Y3 A0A3B3RF51 A0A3B3RHI3 A0A3B3RFR1 A0A3B3REZ2 A0A3B3RF49 M3ZTQ6 A0A158PSU0 A0A0H5SLJ0 A0A3P7DE29 A0A0K0JMA7 A0A3Q2Y913 A0A3Q2Y7L2 A0A3B5PRK5 A0A1D1W8N8 A0A0R3QL57 A0A3B5PQT5 A0A3P6SR81
PDB
6A69
E-value=5.98404e-07,
Score=132
Ontologies
GO
Topology
Subcellular location
Membrane
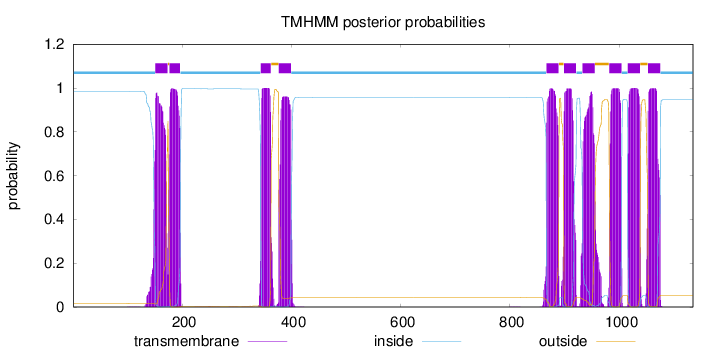
Length:
1135
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
219.21442
Exp number, first 60 AAs:
0
Total prob of N-in:
0.98317
inside
1 - 150
TMhelix
151 - 173
outside
174 - 176
TMhelix
177 - 196
inside
197 - 343
TMhelix
344 - 362
outside
363 - 376
TMhelix
377 - 399
inside
400 - 866
TMhelix
867 - 889
outside
890 - 898
TMhelix
899 - 921
inside
922 - 932
TMhelix
933 - 955
outside
956 - 981
TMhelix
982 - 1004
inside
1005 - 1015
TMhelix
1016 - 1038
outside
1039 - 1052
TMhelix
1053 - 1075
inside
1076 - 1135
Population Genetic Test Statistics
Pi
272.627435
Theta
167.483711
Tajima's D
2.484662
CLR
0.25631
CSRT
0.942402879856007
Interpretation
Uncertain
