Gene
KWMTBOMO15989
Annotation
PREDICTED:_ankyrin-3-like?_partial_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.09 Mitochondrial Reliability : 1.197
Sequence
CDS
ATGGGTGGACGAGCTCACAGCCCACCTGGTATTAAGTGGTTACTGGAGCCCATAGACATCCACAACATAGATCCAAACACAGCATTTTTGCGAGCAGCACGTGCCGGTCAGCTCGACACAGTAATTGAGCTCCTCGATTCGGGCGCTGTCAAAGACATCAACACATGTAATACGAATGGACTGAATGCTCTTCATCTAGCAGCGAAAGATGGTCACATATCAGTGGTCGAGGAGCTGCTCAAACGGGGCGCCACCGTGGACGCTGCAACTAAAAAAGGCAACACAGCCCTACACATAGCATGTCTCGCGGGGCAGGAGGCGGTGGCGAAAGCCCTTCTCGGGGCCGGGGCGAAGGCCGACGCCCAATCCGCAGCAGGCTTCACACCTCTCTACATGGCTGCACAAGAGAATCATGCTGGATGCGTCAAGATGCTGCTTGCCGCTGGAGCCAGTCAAACTCTCGCTACAGAAGATGGTTTCACGCCATTAGCGGTGGCCATGCAGCAAGGACACGACAGGGTAGTCGCGGAACTACTGGAATCCGACACAAGGGGCAAGGTTCGATTGCCGGCGTTACACATCGCGGCCAAGAAAAATGACGTGAAAGCGGCAACTCTGCTGTTAGAGAACGAGCACAACCCGGACGCGTGCTCCAAGTCGGGGTTCACGCCGCTGCACATCGCGGCGCACTACGGGAACGCGGGCGTGGCGCGCGCGCTGCTGGCCGCCGGCGCGGACCCCGCGCGCGCCGCCAAGCACGCCATCACGCCGCTGCACGTGGCCAGCAAGTGGGGCCAGCTCGCCATGCTCGACCTGCTGCTCGAACACGGCGCGAATATAGCAGCAGTGACCCGCGACGGTCTGACCCCGCTACATTGCGCGGCGCGATCCGGACACAGCAATGTAGTGTCGCGGCTGCTGCAACACGGTGCGCCGATCACCAGCAAGACTAAGAATGGACTGACTCCTCTCCACATGTCCGTGCAAGGCGAACACGTGGAGACGGCCAAAGTACTCCTGACAGAGGGCGCGCCCATCGATGACGTCACAGTCGATTACCTCACCGCACTGCATGTCGCGGCTCATTGTGGACACGTCAAGGTGGCTAAACTGTTGTTAGATAGAAACGCGGATGCGAACGCACGAGCGCTGAACGGGTTCACGCCCCTCCACATCGCTTGCAAAAAGAACAGACTCAAAGTCGTCGAGTTGCTTCTTAAATATGGCGCCAGTAAATCGGCGACCACCGAGTCGGGCCTGACGCCTCTCCACGTGGCGTCGTTCATGGGCTGCATGAACATAGCGCTGGTGCTCATAGGGGCGGGCGCGGACGCCGACGCCGCCACGGCCCGCGGGGAGACGCCGCTGCACCTGGCCGCCAGGGCGCACCAGACCGACCTGGTCCGGGTGCTCCTCAGGAATAACGCTAAGGTGGACGCGCGCGCCCGCGAGGAGCAGAGCCCGCTGCACGTGGCGGCGCGGCTGGGGCACGCCGACATCGCGGCGCTGCTGCTGCAGCACGGCGCCGACGTGGCCGCCACCACCAAGGATCGGTACACGCCGCTGCACATCGCCGCCAAAGAAGGCAAAGAAGAAGTAGCGGCTATACTCCTGGACAACAACGCGCCCATAGAAGCGGAGACGCGCAAAGGGTTCACGCCGCTCCACCTGGCGGCCAAGTACGGAGACATCGGCGTGGCGCGGCTGCTGCTGGCCCGCGGGGCGCAGCCCGACGCGCCCGGCAAGAGCCACATCACCCCCCTGCACATGGCCACCTACTACGGACACCCCGACATCGCCTTGCTGCTGTTAGATAAAGGCGCATCTCCGCATTCGTTAGCCAAAAACGGTCACTCGGCTCTGCACATCGCCTGTCGGCACAACCACCCCGATATCGCCTTTGCACTTCTTGAACACGATGCCGATCCCGGGGTAAAATCGAAAGCCGGGTTCACACCTCTACACATGGCGGCACAAGAGGGACACGAGGACTGCGTCGAGATGCTCATCGAGAGAGGAGCTGACGTTAATGTACCTGCAAATAATGGTCTGACTCCGGTGCACCTGGCTGCGTCGGAGGGTCGCACGGGCGTGCTGAAGGCGCTGCTGGCGGCGGGCGGGCAGTGCTCCGCCAAGACGCGCGACGGGTACACGCCGCTGCACGCCGCCGCGCACCACGGTCACCACGCCGCCGCCAGGACGCTCATCGATGCCGACGCTGACGTCACAGCTAAAGCCGCGCACGGGTTCACGCCGCTGCACCAGGCCGCGCAACAAGGACACACGCTTATTATACAGCTGCTGTTGAAACACAACGCCGACCCCAACGCACTGTCAGCCAACGGTCAGACGCCGTGTGCCATCGCGGACCGGCTGGGTTACATCAGCGCGGTGGAGGCGCTGCGCCCCGTCACCGAGAACACGCTCAGTCAGGCCGTCGGGGACACCGGTAAGTGGCGTGCTTCAATAAGACTCCTACCTTGA
Protein
MGGRAHSPPGIKWLLEPIDIHNIDPNTAFLRAARAGQLDTVIELLDSGAVKDINTCNTNGLNALHLAAKDGHISVVEELLKRGATVDAATKKGNTALHIACLAGQEAVAKALLGAGAKADAQSAAGFTPLYMAAQENHAGCVKMLLAAGASQTLATEDGFTPLAVAMQQGHDRVVAELLESDTRGKVRLPALHIAAKKNDVKAATLLLENEHNPDACSKSGFTPLHIAAHYGNAGVARALLAAGADPARAAKHAITPLHVASKWGQLAMLDLLLEHGANIAAVTRDGLTPLHCAARSGHSNVVSRLLQHGAPITSKTKNGLTPLHMSVQGEHVETAKVLLTEGAPIDDVTVDYLTALHVAAHCGHVKVAKLLLDRNADANARALNGFTPLHIACKKNRLKVVELLLKYGASKSATTESGLTPLHVASFMGCMNIALVLIGAGADADAATARGETPLHLAARAHQTDLVRVLLRNNAKVDARAREEQSPLHVAARLGHADIAALLLQHGADVAATTKDRYTPLHIAAKEGKEEVAAILLDNNAPIEAETRKGFTPLHLAAKYGDIGVARLLLARGAQPDAPGKSHITPLHMATYYGHPDIALLLLDKGASPHSLAKNGHSALHIACRHNHPDIAFALLEHDADPGVKSKAGFTPLHMAAQEGHEDCVEMLIERGADVNVPANNGLTPVHLAASEGRTGVLKALLAAGGQCSAKTRDGYTPLHAAAHHGHHAAARTLIDADADVTAKAAHGFTPLHQAAQQGHTLIIQLLLKHNADPNALSANGQTPCAIADRLGYISAVEALRPVTENTLSQAVGDTGKWRASIRLLP
Summary
Uniprot
A0A3S2NQ91
A0A194Q539
A0A2A4KA56
A0A212FLP2
A0A088A5I7
A0A151J9H4
+ More
A0A0M9A139 A0A0C9RX19 A0A154P4Q5 A0A0L7QTB8 A0A139WK59 A0A139WK00 A0A139WKL7 A0A0P6IVZ9 A0A0C9RX27 V9I882 Q16J24 V5GGY4 V9I7C5 V9I6F6 V9I7S8 A0A1B6LXG4 A0A1B6K8F2 A0A1W4WI54 K7IZZ3 A0A1Q3FUC6 A0A1Q3FUA3 W4VRE6 A0A1W4WU08 A0A139WKF6 A0A139WK40 A0A0J7LB10 A0A182W8X3 A0A139WK56 A0A084WG27 A0A182RRB6 A0A139WKB6 A0A182NAV6 E9JCY0 A0A2J7QN88 A0A182R053 A0A182F4Q0 A0A182HRE5 A0A336MI74 A0A1S4HE56 A0A336MM38 A0A0L0CHH3 A0A182TPC3 F5HIS2 A0A336MJL2 A0A336MVZ3 A0A2J7QN90 A0A087ZFH8 A0A2M4CR79 A0A336MJP7 A0A336MMS2 A0A336MI87 A0A1B6L5U9 A0A336MKS6 A0A310SGH9 A0A1W5C970 A0A1B6LJC9 F5HIS1 W5J9Y1 A0A1B6L0M3 A0A2M3YYR0 A0A182QUQ5 A0A182ND69 A0A1B6KJ18 A0A182FT56 A0A1B6C335 A0A182Y4S7 A0A1B6G621 A0A1B6BWC3 A0A182X3C2 A0A158NYG9 A0A1W4WHD6 A0A1W4WTJ8 A0A1I8P656 Q7QKD3 Q16PP6 A0A336MKZ5 A0A182R2C7 A0A182K4G2 A0A195CXE2 A0A182VQ03 A0A182UYY2 A0A151WHJ1 F4WT31 A0A195F701 A0A023F410 N6UGE5 A0A0K8T331 A0A0K8T338 A0A0A9YPD4 A0A0A9Y3T9 A0A0A9Y5E1 A0A0K8T3F5 A0A0A9Y6Z1 A0A0K8T335
A0A0M9A139 A0A0C9RX19 A0A154P4Q5 A0A0L7QTB8 A0A139WK59 A0A139WK00 A0A139WKL7 A0A0P6IVZ9 A0A0C9RX27 V9I882 Q16J24 V5GGY4 V9I7C5 V9I6F6 V9I7S8 A0A1B6LXG4 A0A1B6K8F2 A0A1W4WI54 K7IZZ3 A0A1Q3FUC6 A0A1Q3FUA3 W4VRE6 A0A1W4WU08 A0A139WKF6 A0A139WK40 A0A0J7LB10 A0A182W8X3 A0A139WK56 A0A084WG27 A0A182RRB6 A0A139WKB6 A0A182NAV6 E9JCY0 A0A2J7QN88 A0A182R053 A0A182F4Q0 A0A182HRE5 A0A336MI74 A0A1S4HE56 A0A336MM38 A0A0L0CHH3 A0A182TPC3 F5HIS2 A0A336MJL2 A0A336MVZ3 A0A2J7QN90 A0A087ZFH8 A0A2M4CR79 A0A336MJP7 A0A336MMS2 A0A336MI87 A0A1B6L5U9 A0A336MKS6 A0A310SGH9 A0A1W5C970 A0A1B6LJC9 F5HIS1 W5J9Y1 A0A1B6L0M3 A0A2M3YYR0 A0A182QUQ5 A0A182ND69 A0A1B6KJ18 A0A182FT56 A0A1B6C335 A0A182Y4S7 A0A1B6G621 A0A1B6BWC3 A0A182X3C2 A0A158NYG9 A0A1W4WHD6 A0A1W4WTJ8 A0A1I8P656 Q7QKD3 Q16PP6 A0A336MKZ5 A0A182R2C7 A0A182K4G2 A0A195CXE2 A0A182VQ03 A0A182UYY2 A0A151WHJ1 F4WT31 A0A195F701 A0A023F410 N6UGE5 A0A0K8T331 A0A0K8T338 A0A0A9YPD4 A0A0A9Y3T9 A0A0A9Y5E1 A0A0K8T3F5 A0A0A9Y6Z1 A0A0K8T335
Pubmed
EMBL
RSAL01000012
RVE53499.1
KQ459460
KPJ00653.1
NWSH01000025
PCG80610.1
+ More
AGBW02007724 OWR54663.1 KQ979403 KYN21720.1 KQ435775 KOX75027.1 GBYB01013630 JAG83397.1 KQ434817 KZC06871.1 KQ414747 KOC61883.1 KQ971328 KYB28369.1 KYB28368.1 KYB28367.1 GDUN01000063 JAN95856.1 GBYB01013640 JAG83407.1 JR035606 AEY56539.1 CH478035 EAT34273.1 GALX01005207 JAB63259.1 JR035603 AEY56537.1 JR035605 AEY56538.1 JR035602 AEY56536.1 GEBQ01011642 JAT28335.1 GEBQ01032238 JAT07739.1 GFDL01003846 JAV31199.1 GFDL01003844 JAV31201.1 GANO01003559 JAB56312.1 KQ971332 KYB28285.1 KYB28283.1 LBMM01000066 KMR05097.1 KYB28284.1 ATLV01023434 KE525343 KFB49171.1 KYB28281.1 GL771866 EFZ09225.1 NEVH01013198 PNF30054.1 AXCN02000654 APCN01001695 APCN01001696 UFQT01001345 SSX30122.1 AAAB01008980 SSX30119.1 JRES01000375 KNC31858.1 AAAB01008799 EGK96183.1 SSX30120.1 SSX30118.1 PNF30051.1 GGFL01003220 MBW67398.1 SSX30125.1 SSX30123.1 SSX30124.1 GEBQ01020910 JAT19067.1 SSX30121.1 KQ761592 OAD57374.1 EAA14062.4 GEBQ01016181 JAT23796.1 EGK96182.1 ADMH02001962 ETN60218.1 GEBQ01022757 JAT17220.1 GGFM01000644 MBW21395.1 AXCN02001215 GEBQ01028536 JAT11441.1 GEDC01029524 JAS07774.1 GECZ01011883 JAS57886.1 GEDC01031701 JAS05597.1 ADTU01003819 ADTU01003820 ADTU01003821 ADTU01003822 ADTU01003823 ADTU01003824 ADTU01003825 EAA03765.4 CH477775 EAT36335.1 UFQS01000711 UFQT01000711 SSX06344.1 SSX26698.1 KQ977220 KYN04819.1 KQ983114 KYQ47306.1 GL888331 EGI62644.1 KQ981768 KYN35967.1 GBBI01002507 JAC16205.1 APGK01026876 KB740648 ENN79711.1 GBRD01005882 JAG59939.1 GBRD01005877 JAG59944.1 GBHO01009555 JAG34049.1 GBHO01016755 JAG26849.1 GBHO01016753 JAG26851.1 GBRD01005875 JAG59946.1 GBHO01016751 JAG26853.1 GBRD01005883 JAG59938.1
AGBW02007724 OWR54663.1 KQ979403 KYN21720.1 KQ435775 KOX75027.1 GBYB01013630 JAG83397.1 KQ434817 KZC06871.1 KQ414747 KOC61883.1 KQ971328 KYB28369.1 KYB28368.1 KYB28367.1 GDUN01000063 JAN95856.1 GBYB01013640 JAG83407.1 JR035606 AEY56539.1 CH478035 EAT34273.1 GALX01005207 JAB63259.1 JR035603 AEY56537.1 JR035605 AEY56538.1 JR035602 AEY56536.1 GEBQ01011642 JAT28335.1 GEBQ01032238 JAT07739.1 GFDL01003846 JAV31199.1 GFDL01003844 JAV31201.1 GANO01003559 JAB56312.1 KQ971332 KYB28285.1 KYB28283.1 LBMM01000066 KMR05097.1 KYB28284.1 ATLV01023434 KE525343 KFB49171.1 KYB28281.1 GL771866 EFZ09225.1 NEVH01013198 PNF30054.1 AXCN02000654 APCN01001695 APCN01001696 UFQT01001345 SSX30122.1 AAAB01008980 SSX30119.1 JRES01000375 KNC31858.1 AAAB01008799 EGK96183.1 SSX30120.1 SSX30118.1 PNF30051.1 GGFL01003220 MBW67398.1 SSX30125.1 SSX30123.1 SSX30124.1 GEBQ01020910 JAT19067.1 SSX30121.1 KQ761592 OAD57374.1 EAA14062.4 GEBQ01016181 JAT23796.1 EGK96182.1 ADMH02001962 ETN60218.1 GEBQ01022757 JAT17220.1 GGFM01000644 MBW21395.1 AXCN02001215 GEBQ01028536 JAT11441.1 GEDC01029524 JAS07774.1 GECZ01011883 JAS57886.1 GEDC01031701 JAS05597.1 ADTU01003819 ADTU01003820 ADTU01003821 ADTU01003822 ADTU01003823 ADTU01003824 ADTU01003825 EAA03765.4 CH477775 EAT36335.1 UFQS01000711 UFQT01000711 SSX06344.1 SSX26698.1 KQ977220 KYN04819.1 KQ983114 KYQ47306.1 GL888331 EGI62644.1 KQ981768 KYN35967.1 GBBI01002507 JAC16205.1 APGK01026876 KB740648 ENN79711.1 GBRD01005882 JAG59939.1 GBRD01005877 JAG59944.1 GBHO01009555 JAG34049.1 GBHO01016755 JAG26849.1 GBHO01016753 JAG26851.1 GBRD01005875 JAG59946.1 GBHO01016751 JAG26853.1 GBRD01005883 JAG59938.1
Proteomes
UP000283053
UP000053268
UP000218220
UP000007151
UP000005203
UP000078492
+ More
UP000053105 UP000076502 UP000053825 UP000007266 UP000008820 UP000192223 UP000002358 UP000036403 UP000075920 UP000030765 UP000075900 UP000075884 UP000235965 UP000075886 UP000069272 UP000075840 UP000037069 UP000075902 UP000007062 UP000000673 UP000076408 UP000076407 UP000005205 UP000095300 UP000075881 UP000078542 UP000075903 UP000075809 UP000007755 UP000078541 UP000019118
UP000053105 UP000076502 UP000053825 UP000007266 UP000008820 UP000192223 UP000002358 UP000036403 UP000075920 UP000030765 UP000075900 UP000075884 UP000235965 UP000075886 UP000069272 UP000075840 UP000037069 UP000075902 UP000007062 UP000000673 UP000076408 UP000076407 UP000005205 UP000095300 UP000075881 UP000078542 UP000075903 UP000075809 UP000007755 UP000078541 UP000019118
Pfam
Interpro
IPR040745
Ankyrin_UPA
+ More
IPR000906 ZU5_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR002110 Ankyrin_rpt
IPR011029 DEATH-like_dom_sf
IPR000074 ApoA_E
IPR000488 Death_domain
IPR009091 RCC1/BLIP-II
IPR011333 SKP1/BTB/POZ_sf
IPR000210 BTB/POZ_dom
IPR000408 Reg_chr_condens
IPR000906 ZU5_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR002110 Ankyrin_rpt
IPR011029 DEATH-like_dom_sf
IPR000074 ApoA_E
IPR000488 Death_domain
IPR009091 RCC1/BLIP-II
IPR011333 SKP1/BTB/POZ_sf
IPR000210 BTB/POZ_dom
IPR000408 Reg_chr_condens
Gene 3D
CDD
ProteinModelPortal
A0A3S2NQ91
A0A194Q539
A0A2A4KA56
A0A212FLP2
A0A088A5I7
A0A151J9H4
+ More
A0A0M9A139 A0A0C9RX19 A0A154P4Q5 A0A0L7QTB8 A0A139WK59 A0A139WK00 A0A139WKL7 A0A0P6IVZ9 A0A0C9RX27 V9I882 Q16J24 V5GGY4 V9I7C5 V9I6F6 V9I7S8 A0A1B6LXG4 A0A1B6K8F2 A0A1W4WI54 K7IZZ3 A0A1Q3FUC6 A0A1Q3FUA3 W4VRE6 A0A1W4WU08 A0A139WKF6 A0A139WK40 A0A0J7LB10 A0A182W8X3 A0A139WK56 A0A084WG27 A0A182RRB6 A0A139WKB6 A0A182NAV6 E9JCY0 A0A2J7QN88 A0A182R053 A0A182F4Q0 A0A182HRE5 A0A336MI74 A0A1S4HE56 A0A336MM38 A0A0L0CHH3 A0A182TPC3 F5HIS2 A0A336MJL2 A0A336MVZ3 A0A2J7QN90 A0A087ZFH8 A0A2M4CR79 A0A336MJP7 A0A336MMS2 A0A336MI87 A0A1B6L5U9 A0A336MKS6 A0A310SGH9 A0A1W5C970 A0A1B6LJC9 F5HIS1 W5J9Y1 A0A1B6L0M3 A0A2M3YYR0 A0A182QUQ5 A0A182ND69 A0A1B6KJ18 A0A182FT56 A0A1B6C335 A0A182Y4S7 A0A1B6G621 A0A1B6BWC3 A0A182X3C2 A0A158NYG9 A0A1W4WHD6 A0A1W4WTJ8 A0A1I8P656 Q7QKD3 Q16PP6 A0A336MKZ5 A0A182R2C7 A0A182K4G2 A0A195CXE2 A0A182VQ03 A0A182UYY2 A0A151WHJ1 F4WT31 A0A195F701 A0A023F410 N6UGE5 A0A0K8T331 A0A0K8T338 A0A0A9YPD4 A0A0A9Y3T9 A0A0A9Y5E1 A0A0K8T3F5 A0A0A9Y6Z1 A0A0K8T335
A0A0M9A139 A0A0C9RX19 A0A154P4Q5 A0A0L7QTB8 A0A139WK59 A0A139WK00 A0A139WKL7 A0A0P6IVZ9 A0A0C9RX27 V9I882 Q16J24 V5GGY4 V9I7C5 V9I6F6 V9I7S8 A0A1B6LXG4 A0A1B6K8F2 A0A1W4WI54 K7IZZ3 A0A1Q3FUC6 A0A1Q3FUA3 W4VRE6 A0A1W4WU08 A0A139WKF6 A0A139WK40 A0A0J7LB10 A0A182W8X3 A0A139WK56 A0A084WG27 A0A182RRB6 A0A139WKB6 A0A182NAV6 E9JCY0 A0A2J7QN88 A0A182R053 A0A182F4Q0 A0A182HRE5 A0A336MI74 A0A1S4HE56 A0A336MM38 A0A0L0CHH3 A0A182TPC3 F5HIS2 A0A336MJL2 A0A336MVZ3 A0A2J7QN90 A0A087ZFH8 A0A2M4CR79 A0A336MJP7 A0A336MMS2 A0A336MI87 A0A1B6L5U9 A0A336MKS6 A0A310SGH9 A0A1W5C970 A0A1B6LJC9 F5HIS1 W5J9Y1 A0A1B6L0M3 A0A2M3YYR0 A0A182QUQ5 A0A182ND69 A0A1B6KJ18 A0A182FT56 A0A1B6C335 A0A182Y4S7 A0A1B6G621 A0A1B6BWC3 A0A182X3C2 A0A158NYG9 A0A1W4WHD6 A0A1W4WTJ8 A0A1I8P656 Q7QKD3 Q16PP6 A0A336MKZ5 A0A182R2C7 A0A182K4G2 A0A195CXE2 A0A182VQ03 A0A182UYY2 A0A151WHJ1 F4WT31 A0A195F701 A0A023F410 N6UGE5 A0A0K8T331 A0A0K8T338 A0A0A9YPD4 A0A0A9Y3T9 A0A0A9Y5E1 A0A0K8T3F5 A0A0A9Y6Z1 A0A0K8T335
PDB
4RLV
E-value=0,
Score=2043
Ontologies
GO
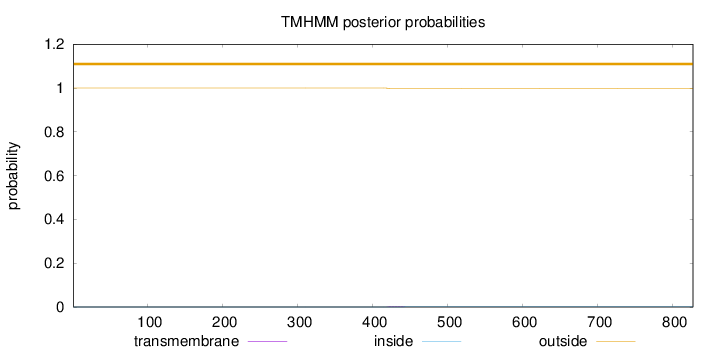
Topology
Length:
827
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0509800000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00032
outside
1 - 827
Population Genetic Test Statistics
Pi
158.369711
Theta
150.832803
Tajima's D
0.215861
CLR
104.217548
CSRT
0.432528373581321
Interpretation
Uncertain
