Pre Gene Modal
BGIBMGA005727
Annotation
PREDICTED:_pyrroline-5-carboxylate_reductase-like_isoform_X1_[Papilio_xuthus]
Full name
Pyrroline-5-carboxylate reductase
Location in the cell
Mitochondrial Reliability : 1.937
Sequence
CDS
ATGCTGAAATTACCATTAATAGAAGCTATGAAGATCGGATTTATAGGCGGCGGCAGGCTTGCGTTTGCTTTGGCGAACGGCTTTATTTCGGCGGGTCTGGCAAAACCCGACGAAATCACAGCTAGCTGTCATCCTTCCGATGTTGCCAGCGCCAAAGCTTTTCAGGGCTTGGGAGCTACAGCGTTATTTGAAAATATATCCGTGGTGGAGAGGTCTGAAGTGGTGATAGTATCTGTCAAGCCCGATGTCGTGGTACCCGCTCTTAAAGAGATAAAAAACCTGCCTTCGGCGAAGAATAAACTCTTCATTTCCGTTGCCATGGGAATTACTATCGCTACAGTCGAATCGAATTTGCCTTCTGAGGCTCGTGTGATTCGTGTAATGCCAAACACTCCGGCGTTAGTGAAAGAAGGCGCAGCTGCCTTTAGTCGTGGTACTAAAGCCACAGCGGAAGACGCACAACTGACGTCACAGCTCTTCAAGGCGGTGGGTACCTGCGATGAGGTACCAGAGTACCAAATGGACGCTATAACCGCCCTTAGTGGAAGTGGACCCGCTTACGTTTACATGCTAATAGAATCTCTAGCTGATGGTGGAGTTCGTTGTGGCTTACCTCGCGACCTCGCACTTCGCCTGGCCGCCCAAACCACCCTGGGTTCCGCAGCCATGGTCAAAACTGGTGATCATCCAGCCATGTTGAAAGACAATGTCACCAGTCCCGCAGGGTCTACCGCTGAGGGAACATATCACTTGGAGAAAAACGGTTTCCGTTCTGCCATAATCGGTGCAGTGTCTGCGGCAGTAGATAGATGTAAGATTGTAAAAAAACAATTGAACGATTAG
Protein
MLKLPLIEAMKIGFIGGGRLAFALANGFISAGLAKPDEITASCHPSDVASAKAFQGLGATALFENISVVERSEVVIVSVKPDVVVPALKEIKNLPSAKNKLFISVAMGITIATVESNLPSEARVIRVMPNTPALVKEGAAAFSRGTKATAEDAQLTSQLFKAVGTCDEVPEYQMDAITALSGSGPAYVYMLIESLADGGVRCGLPRDLALRLAAQTTLGSAAMVKTGDHPAMLKDNVTSPAGSTAEGTYHLEKNGFRSAIIGAVSAAVDRCKIVKKQLND
Summary
Catalytic Activity
L-proline + NADP(+) = 1-pyrroline-5-carboxylate + 2 H(+) + NADPH
Similarity
Belongs to the pyrroline-5-carboxylate reductase family.
Feature
chain Pyrroline-5-carboxylate reductase
Uniprot
H9J885
A0A0N1PJ82
A0A194Q6L9
A0A2A4KA00
A0A2W1BFW2
A0A2A4KAD6
+ More
A0A2H1VGZ2 A0A212EIV7 D6WHI9 A0A1B6KM20 A0A1B6I3B3 J9WM64 A0A1J1HJ17 A0A1A9WNS5 A0A1L8E2K6 J3JUU5 G9FQ76 A0A1A9UEM6 A0A1B0C6T3 A0A1A9X9X1 A0A1A9YUI0 U5EUM2 A0A1L8EFP3 B4JTU7 A0A1S4FFD3 Q173T1 Q2KQA0 Q29A90 A0A1B6E9L7 A0A1L8EBY5 A0A0L0C7U0 A0A182N1U1 A0A182GUZ7 A0A182J885 E0VU70 A0A182R2B9 A0A0K8U985 A0A034WHX1 A0A0C9RP24 A0A182U1W0 B3LVQ0 A0A3B0K4U7 A0A182VAQ4 A0A182KNJ5 Q7PPH9 A0A182HQR1 A0A146KKA3 N6U3D7 A0A0A9WR31 A0A1I8PIX4 A0A182WM59 Q9VEJ3 A0A182FLD0 A0A2M3Z7E7 A0A2M3Z7F8 B3P3D2 B4PKF1 A0A0M5JCD7 A0A1Y1KTM8 A0A1W4V516 Q0QHK5 W5JE89 B4QVP6 B4LY47 B4IBG7 A0A2M4AQV9 B4K7N1 T1PG15 A0A3L8DJ27 A0A2M4BWS7 B4G2L5 K7J149 A0A232FIG1 B4N8G5 A0A0P4VK41 A0A224XHV6 A0A0A1XKR3 A0A0K8VNU6 W8AYB1 A0A2S2QXU0 B0VZ26 A0A195F9T8 A0A034WGW1 C4WS23 E1ZX14 A0A1Q3FNK1 A0A0L7QQL9 A0A0J7L134 A0A182S9T1 A0A170YTU3 A0A0C9QTC7 A0A2H8TXC9 T1HIQ9 A0A154PFY4 A0A158NP73 F4WF31 A0A0L7LI33 A0A195DYH3
A0A2H1VGZ2 A0A212EIV7 D6WHI9 A0A1B6KM20 A0A1B6I3B3 J9WM64 A0A1J1HJ17 A0A1A9WNS5 A0A1L8E2K6 J3JUU5 G9FQ76 A0A1A9UEM6 A0A1B0C6T3 A0A1A9X9X1 A0A1A9YUI0 U5EUM2 A0A1L8EFP3 B4JTU7 A0A1S4FFD3 Q173T1 Q2KQA0 Q29A90 A0A1B6E9L7 A0A1L8EBY5 A0A0L0C7U0 A0A182N1U1 A0A182GUZ7 A0A182J885 E0VU70 A0A182R2B9 A0A0K8U985 A0A034WHX1 A0A0C9RP24 A0A182U1W0 B3LVQ0 A0A3B0K4U7 A0A182VAQ4 A0A182KNJ5 Q7PPH9 A0A182HQR1 A0A146KKA3 N6U3D7 A0A0A9WR31 A0A1I8PIX4 A0A182WM59 Q9VEJ3 A0A182FLD0 A0A2M3Z7E7 A0A2M3Z7F8 B3P3D2 B4PKF1 A0A0M5JCD7 A0A1Y1KTM8 A0A1W4V516 Q0QHK5 W5JE89 B4QVP6 B4LY47 B4IBG7 A0A2M4AQV9 B4K7N1 T1PG15 A0A3L8DJ27 A0A2M4BWS7 B4G2L5 K7J149 A0A232FIG1 B4N8G5 A0A0P4VK41 A0A224XHV6 A0A0A1XKR3 A0A0K8VNU6 W8AYB1 A0A2S2QXU0 B0VZ26 A0A195F9T8 A0A034WGW1 C4WS23 E1ZX14 A0A1Q3FNK1 A0A0L7QQL9 A0A0J7L134 A0A182S9T1 A0A170YTU3 A0A0C9QTC7 A0A2H8TXC9 T1HIQ9 A0A154PFY4 A0A158NP73 F4WF31 A0A0L7LI33 A0A195DYH3
EC Number
1.5.1.2
Pubmed
19121390
26354079
28756777
22118469
18362917
19820115
+ More
22972362 22516182 17994087 17510324 15804581 15632085 26108605 26483478 20566863 25348373 20966253 12364791 14747013 17210077 26823975 23537049 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 28004739 20920257 23761445 25315136 30249741 20075255 28648823 27129103 25830018 24495485 20798317 21347285 21719571 26227816
22972362 22516182 17994087 17510324 15804581 15632085 26108605 26483478 20566863 25348373 20966253 12364791 14747013 17210077 26823975 23537049 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 28004739 20920257 23761445 25315136 30249741 20075255 28648823 27129103 25830018 24495485 20798317 21347285 21719571 26227816
EMBL
BABH01038495
KQ459890
KPJ19552.1
KQ459460
KPJ00650.1
NWSH01000025
+ More
PCG80613.1 KZ150219 PZC72117.1 PCG80612.1 ODYU01002517 SOQ40113.1 AGBW02014566 OWR41400.1 KQ971321 EFA00672.1 GEBQ01027510 JAT12467.1 GECU01026341 GECU01003200 JAS81365.1 JAT04507.1 JX462668 AFS17324.1 CVRI01000006 CRK88005.1 GFDF01001170 JAV12914.1 BT127010 AEE61972.1 JN187430 AET80386.2 JXJN01026961 CCAG010011557 GANO01002260 JAB57611.1 GFDG01001291 JAV17508.1 CH916374 EDV91526.1 CH477417 EAT41304.1 AY623403 AAV31912.1 CM000070 EAL27460.2 GEDC01003386 GEDC01002744 JAS33912.1 JAS34554.1 GFDG01002531 JAV16268.1 JRES01000789 KNC28326.1 JXUM01019405 KQ560541 KXJ81941.1 DS235780 EEB16926.1 GDHF01029161 JAI23153.1 GAKP01005579 JAC53373.1 GBYB01010185 JAG79952.1 CH902617 EDV43674.2 OUUW01000005 SPP80646.1 AAAB01008948 EAA10379.5 APCN01004681 GDHC01021616 JAP97012.1 APGK01041389 KB740993 ENN76080.1 GBHO01033370 GBHO01033362 JAG10234.1 JAG10242.1 AE014297 AY071558 BT010232 AAF55428.1 AAL49180.1 AAQ23550.1 GGFM01003691 MBW24442.1 GGFM01003690 MBW24441.1 CH954181 EDV48712.1 CM000160 EDW95790.1 CP012526 ALC46294.1 GEZM01074059 JAV64684.1 DQ374014 ABG77992.1 ADMH02001638 ETN61658.1 CM000364 EDX12559.1 CH940650 EDW67935.1 CH480827 EDW44725.1 GGFK01009667 MBW42988.1 CH933806 EDW15375.1 KA647701 AFP62330.1 QOIP01000007 RLU20434.1 GGFJ01008323 MBW57464.1 CH479179 EDW24060.1 NNAY01000180 OXU30248.1 CH964232 EDW81416.1 GDKW01001932 JAI54663.1 GFTR01004359 JAW12067.1 GBXI01003154 JAD11138.1 GDHF01011750 JAI40564.1 GAMC01016717 GAMC01016716 JAB89838.1 GGMS01013373 MBY82576.1 DS231813 EDS25746.1 KQ981727 KYN36814.1 GAKP01005577 JAC53375.1 ABLF02032883 ABLF02032887 ABLF02045861 AK340016 BAH70693.1 GL434948 EFN74284.1 GFDL01005963 JAV29082.1 KQ414786 KOC60923.1 LBMM01001448 KMQ96293.1 GEMB01002968 JAS00235.1 GBYB01006964 JAG76731.1 GFXV01007091 MBW18896.1 ACPB03009651 KQ434896 KZC10751.1 ADTU01022267 ADTU01022268 GL888115 EGI67082.1 JTDY01001021 KOB75112.1 KQ980066 KYN17928.1
PCG80613.1 KZ150219 PZC72117.1 PCG80612.1 ODYU01002517 SOQ40113.1 AGBW02014566 OWR41400.1 KQ971321 EFA00672.1 GEBQ01027510 JAT12467.1 GECU01026341 GECU01003200 JAS81365.1 JAT04507.1 JX462668 AFS17324.1 CVRI01000006 CRK88005.1 GFDF01001170 JAV12914.1 BT127010 AEE61972.1 JN187430 AET80386.2 JXJN01026961 CCAG010011557 GANO01002260 JAB57611.1 GFDG01001291 JAV17508.1 CH916374 EDV91526.1 CH477417 EAT41304.1 AY623403 AAV31912.1 CM000070 EAL27460.2 GEDC01003386 GEDC01002744 JAS33912.1 JAS34554.1 GFDG01002531 JAV16268.1 JRES01000789 KNC28326.1 JXUM01019405 KQ560541 KXJ81941.1 DS235780 EEB16926.1 GDHF01029161 JAI23153.1 GAKP01005579 JAC53373.1 GBYB01010185 JAG79952.1 CH902617 EDV43674.2 OUUW01000005 SPP80646.1 AAAB01008948 EAA10379.5 APCN01004681 GDHC01021616 JAP97012.1 APGK01041389 KB740993 ENN76080.1 GBHO01033370 GBHO01033362 JAG10234.1 JAG10242.1 AE014297 AY071558 BT010232 AAF55428.1 AAL49180.1 AAQ23550.1 GGFM01003691 MBW24442.1 GGFM01003690 MBW24441.1 CH954181 EDV48712.1 CM000160 EDW95790.1 CP012526 ALC46294.1 GEZM01074059 JAV64684.1 DQ374014 ABG77992.1 ADMH02001638 ETN61658.1 CM000364 EDX12559.1 CH940650 EDW67935.1 CH480827 EDW44725.1 GGFK01009667 MBW42988.1 CH933806 EDW15375.1 KA647701 AFP62330.1 QOIP01000007 RLU20434.1 GGFJ01008323 MBW57464.1 CH479179 EDW24060.1 NNAY01000180 OXU30248.1 CH964232 EDW81416.1 GDKW01001932 JAI54663.1 GFTR01004359 JAW12067.1 GBXI01003154 JAD11138.1 GDHF01011750 JAI40564.1 GAMC01016717 GAMC01016716 JAB89838.1 GGMS01013373 MBY82576.1 DS231813 EDS25746.1 KQ981727 KYN36814.1 GAKP01005577 JAC53375.1 ABLF02032883 ABLF02032887 ABLF02045861 AK340016 BAH70693.1 GL434948 EFN74284.1 GFDL01005963 JAV29082.1 KQ414786 KOC60923.1 LBMM01001448 KMQ96293.1 GEMB01002968 JAS00235.1 GBYB01006964 JAG76731.1 GFXV01007091 MBW18896.1 ACPB03009651 KQ434896 KZC10751.1 ADTU01022267 ADTU01022268 GL888115 EGI67082.1 JTDY01001021 KOB75112.1 KQ980066 KYN17928.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000007266
+ More
UP000183832 UP000091820 UP000078200 UP000092460 UP000092443 UP000092444 UP000001070 UP000008820 UP000001819 UP000037069 UP000075884 UP000069940 UP000249989 UP000075880 UP000009046 UP000075900 UP000075902 UP000007801 UP000268350 UP000075903 UP000075882 UP000007062 UP000075840 UP000019118 UP000095300 UP000075920 UP000000803 UP000069272 UP000008711 UP000002282 UP000092553 UP000192221 UP000000673 UP000000304 UP000008792 UP000001292 UP000009192 UP000095301 UP000279307 UP000008744 UP000002358 UP000215335 UP000007798 UP000002320 UP000078541 UP000007819 UP000000311 UP000053825 UP000036403 UP000075901 UP000015103 UP000076502 UP000005205 UP000007755 UP000037510 UP000078492
UP000183832 UP000091820 UP000078200 UP000092460 UP000092443 UP000092444 UP000001070 UP000008820 UP000001819 UP000037069 UP000075884 UP000069940 UP000249989 UP000075880 UP000009046 UP000075900 UP000075902 UP000007801 UP000268350 UP000075903 UP000075882 UP000007062 UP000075840 UP000019118 UP000095300 UP000075920 UP000000803 UP000069272 UP000008711 UP000002282 UP000092553 UP000192221 UP000000673 UP000000304 UP000008792 UP000001292 UP000009192 UP000095301 UP000279307 UP000008744 UP000002358 UP000215335 UP000007798 UP000002320 UP000078541 UP000007819 UP000000311 UP000053825 UP000036403 UP000075901 UP000015103 UP000076502 UP000005205 UP000007755 UP000037510 UP000078492
Interpro
ProteinModelPortal
H9J885
A0A0N1PJ82
A0A194Q6L9
A0A2A4KA00
A0A2W1BFW2
A0A2A4KAD6
+ More
A0A2H1VGZ2 A0A212EIV7 D6WHI9 A0A1B6KM20 A0A1B6I3B3 J9WM64 A0A1J1HJ17 A0A1A9WNS5 A0A1L8E2K6 J3JUU5 G9FQ76 A0A1A9UEM6 A0A1B0C6T3 A0A1A9X9X1 A0A1A9YUI0 U5EUM2 A0A1L8EFP3 B4JTU7 A0A1S4FFD3 Q173T1 Q2KQA0 Q29A90 A0A1B6E9L7 A0A1L8EBY5 A0A0L0C7U0 A0A182N1U1 A0A182GUZ7 A0A182J885 E0VU70 A0A182R2B9 A0A0K8U985 A0A034WHX1 A0A0C9RP24 A0A182U1W0 B3LVQ0 A0A3B0K4U7 A0A182VAQ4 A0A182KNJ5 Q7PPH9 A0A182HQR1 A0A146KKA3 N6U3D7 A0A0A9WR31 A0A1I8PIX4 A0A182WM59 Q9VEJ3 A0A182FLD0 A0A2M3Z7E7 A0A2M3Z7F8 B3P3D2 B4PKF1 A0A0M5JCD7 A0A1Y1KTM8 A0A1W4V516 Q0QHK5 W5JE89 B4QVP6 B4LY47 B4IBG7 A0A2M4AQV9 B4K7N1 T1PG15 A0A3L8DJ27 A0A2M4BWS7 B4G2L5 K7J149 A0A232FIG1 B4N8G5 A0A0P4VK41 A0A224XHV6 A0A0A1XKR3 A0A0K8VNU6 W8AYB1 A0A2S2QXU0 B0VZ26 A0A195F9T8 A0A034WGW1 C4WS23 E1ZX14 A0A1Q3FNK1 A0A0L7QQL9 A0A0J7L134 A0A182S9T1 A0A170YTU3 A0A0C9QTC7 A0A2H8TXC9 T1HIQ9 A0A154PFY4 A0A158NP73 F4WF31 A0A0L7LI33 A0A195DYH3
A0A2H1VGZ2 A0A212EIV7 D6WHI9 A0A1B6KM20 A0A1B6I3B3 J9WM64 A0A1J1HJ17 A0A1A9WNS5 A0A1L8E2K6 J3JUU5 G9FQ76 A0A1A9UEM6 A0A1B0C6T3 A0A1A9X9X1 A0A1A9YUI0 U5EUM2 A0A1L8EFP3 B4JTU7 A0A1S4FFD3 Q173T1 Q2KQA0 Q29A90 A0A1B6E9L7 A0A1L8EBY5 A0A0L0C7U0 A0A182N1U1 A0A182GUZ7 A0A182J885 E0VU70 A0A182R2B9 A0A0K8U985 A0A034WHX1 A0A0C9RP24 A0A182U1W0 B3LVQ0 A0A3B0K4U7 A0A182VAQ4 A0A182KNJ5 Q7PPH9 A0A182HQR1 A0A146KKA3 N6U3D7 A0A0A9WR31 A0A1I8PIX4 A0A182WM59 Q9VEJ3 A0A182FLD0 A0A2M3Z7E7 A0A2M3Z7F8 B3P3D2 B4PKF1 A0A0M5JCD7 A0A1Y1KTM8 A0A1W4V516 Q0QHK5 W5JE89 B4QVP6 B4LY47 B4IBG7 A0A2M4AQV9 B4K7N1 T1PG15 A0A3L8DJ27 A0A2M4BWS7 B4G2L5 K7J149 A0A232FIG1 B4N8G5 A0A0P4VK41 A0A224XHV6 A0A0A1XKR3 A0A0K8VNU6 W8AYB1 A0A2S2QXU0 B0VZ26 A0A195F9T8 A0A034WGW1 C4WS23 E1ZX14 A0A1Q3FNK1 A0A0L7QQL9 A0A0J7L134 A0A182S9T1 A0A170YTU3 A0A0C9QTC7 A0A2H8TXC9 T1HIQ9 A0A154PFY4 A0A158NP73 F4WF31 A0A0L7LI33 A0A195DYH3
PDB
5UAX
E-value=7.6763e-47,
Score=470
Ontologies
PATHWAY
GO
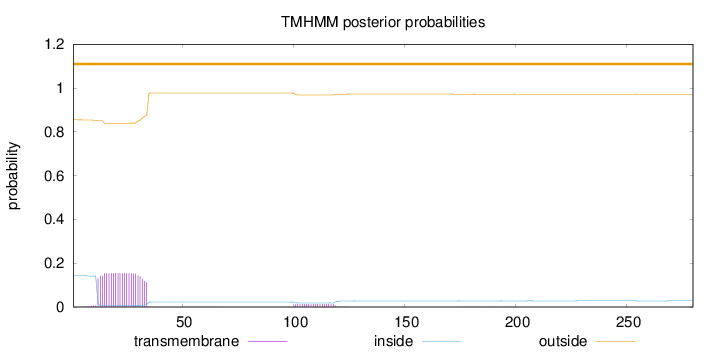
Topology
Length:
280
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.66149
Exp number, first 60 AAs:
3.39842
Total prob of N-in:
0.14528
outside
1 - 280
Population Genetic Test Statistics
Pi
219.513481
Theta
151.788989
Tajima's D
1.438328
CLR
0.164585
CSRT
0.769561521923904
Interpretation
Uncertain
