Gene
KWMTBOMO15985 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005736
Annotation
PREDICTED:_fibulin-1-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.792
Sequence
CDS
ATGAAGTTCATAGTAGTGATTTTTACGTTTTTATTCTCGACCGTCAATGCTTTATCATCGGAAGAGACATTGGCAGTGACAGATTCATGTTGCACCCACGCGGAAGGCGCTGCGTTGCTGTCCACGGACTGTTCAGCTATTCAGCCGCCTGAAGACATAGATGCTGAGCAATTGCCTACCTGTATAACGGCGCTAAAGGCATGCTGCGAGAAACAGCACAAGGAAAAGGACGAATGCTCTGCCGGGGTAAAGTTGGCGAGTTCGAAGAGATGTAACAGCATCGCTACCAACATCGGACAGGCCTGCTGTGAGGAGTGTTCTATTGGTCGCACCATTGGAAAGACTCAAGGATTGGAAGGTTGCGGTACGCCAGTTAATGAGGAGTACAATAGTCCATCGACGATTCTACGCAAGAAAGCTTTTCATCAGTGTTGTTCGGCCGCTGCAAATGAGCCAGAGACAACAGAAAAAGTTGAAACGACAACGCTACCATCCACAACTACATCGACAGCAAAGCCCAAGGTCAAATGCACTAAAGATTCCTGTGCGCAAATCTGTGAGGTCGAGCAAGAGACCATAAGATGTTCGTGCCGTGATGGATTTAGGCTGCATAGCGATAAAAAGTCGTGCAAAGATGTCAACGAATGCGCCGAAGCCGATGATGACATTTGCACAGAAGAAGGATTCGTCTGTCACAACACTCCCGGCTCGTACAAGTGTGTTCCTCTCAAGCCGACAACCAAACGCGACGTGGGATCAAATTGCCCGCCGGGATTCAAAAAGAATGTTCTTAATCAAGTCTGTGATGATATAAATGAATGTCAGCTACCACGACCGCCGTGTCCGAAATACCTTTGCGAGAATACGATAGGAGGCTACAAATGCGCTGGAAAACCGGGCAAGCCAGTGATAGAGTCGGCTCCTGGAGCACCCGAAGTGACGACGCCTGTGGCTCCAGTAAGAAATGAAATTTGCCCTCCAGGCTTCAGAGCTGGACCAGACGACGAATGTATCGACATCGACGAATGCGAGGAGAGTTTGGATGACTGTCAAAGACTTTCTCAACACTGTATAAACACTCATGGTAGTTTCTTCTGTCAAGATCACGTTTCCAAGAGGTGTGCTCCAGGGTTTAAGGTTAATACCTCTACAGGAATTTGTGAAGATATCGATGAATGTGAAGAAAGCACTGAAGTGTGTAAGAGGACTGAAATATGCGTTAATTTACCCGGTGCATACAATTGCAAATCAAAGATCAGCTCTCTTCCCAAACTCGCCAAAACCAAGTGTCAAGAAGGAATGAGACTGCGGCCAGGTGGAACGGTTTGCGAAGATATCGACGAGTGTCGCGAAGGAACGCATCTTTGTGATGAATTTCAAAATTGTATCAATACTTACGGCGCGCATGAGTGTCGCTGCAAGAATGGTTTCGAGTTAGATTCCCTTTTGGAATCCTGTGTGGACATCGACGAGTGTGCGTTGAAGCTAGACAACTGCGGTCCACGCCTGCACTGTCTCAATGTTCTTGGTTCATTTACGTGTACTTATCGCCAACCATCCACTACCTCCACCACACCGGTACCGCCGCTATATGAATACGAATATTACGATTCGGAGGATGACAATGAAACTGCTCCAGAAATCGCAGCAACAACATCAACTACCAGACCAACCTATCCAACTCAAAGAACAACGACATCAACACAGAGAACAACAACATCAACACAGAGAACAACAACATCTACACCAAGAACAACCACATCGACCCAAAGAACAACTACTTCAACTACTAAGCCTACAACATTAACAACACGACGATCTCGCCCATACTATCCATATTATGATAGACCAAGATATACGACTTCAACAAAAAGACCTTCATACCCGTATAATCGTAGACCGAGCACTCCTAGTCGGGTAGAACCAACCACACCAAAAAAACCAGAAGTTACATCGAGCACAGAAGTTTCTCGACGACCAGACTACAATCCCAGTAGATCAGATTACACACCGAGAAGACCGGAAATAACAACGCCGATTAAACCAGAAACTACTACAGAGGTGTTTCGAAGACCTACTCATAGCCCTAGTAGACCAGACAATTCTAGAAGACCTTCGCCGTTTAGACCTGGATATGTCACGGGTATGACAAGGCCAAAAGACCCTATAACTATTGAAAAAGAAAGAGAAGATGATGGCGGTATCGAACTGGATACAAACACAATTCCTAAAGAAACTTGGACAAATGTTATTGGAAGGGACAAGCCAGAAGCTACCGATACACCAGCGTTTGACCCTAACGAAGTACATTGTTTAAACGGATATGAAAAAGACTTCTTTGGCAATTGTGTTGATATAAATGAATGTGAAAGAAGTCATATCTGCAGTAACTTAGAGGACTGCATAAATAAAGCAGGCGGCTATCGCTGCGACTGTATTGCCGGCTTTAAAAGAAACCAGCAAGGATGGTGCATACCCATTCCACCTCCCACTACAACCAGCACTACTTCAACAACGACCAGTACTACCACTCCAAGGACTATTTCTGTATCTTTACCAGAGTGGCAGTATACTCCACCTCGGTCCAGAGGCAGATATCCATCGATTGCTTGTGAACTGGGATACACCTATGACACGAAGTATGGAAAGTGTGTTGATATCGACGAATGCGCGACTAACAAAGCAACGTGCTCTCGAAATGAGGAGTGTGTTAACATCGACGGAGGTTTCCGCTGTGTGTGTGGATGGCGATGCCGAGCTGAAAACGAAAAACCTACCGCAGATCCGTCTTTAGTGCCGGCTTCACCCATATCACCGATTGCCCCACACCCACCACCTCCGTCGTACTCGCCGTCTGAGCCCCGCTACCCCGACAATACCAACGTGATCACGGTGGGTGCGCAATACGGTCAGCGCGGGCCCTGGTCTCTGAGACCTAGCTTCACCCGCTTCTCGGACCGTGGAGCTGTGCCGTGCTCATCGTACTCCCGATGCAAGAACACAGTTGGTTCGTACCGGTGCGAATGCGACGAAGGCTTCCGCAATGCAGCCTCGAATGACAAGGTCTGCGTGGACGTAGACGAGTGCAGCGAGAGTCCGAACGTCTGCCAGCACGGATGCGCCAACACATGGGGCGGATATCGGTGCTACTGCAAGCGGGGCTATCGAGTGGGCAACGATAATAGGACATGCGTGGACGTAAACGAATGCGAAGAGTGGACGTCTGTCCGTATTAGAGGGAAATTGTGCGCTGGCGAATGTGTCAATGAGCCAGGGAGCTATCGATGCGCCTGCCCTGCTGGCTACAGGCTATCTGAAGACAACAGAAGTTGTATAGACATAGACGAATGCGAGACGGGAGAAGCTCCGTGTGCCCGCACGTCAGACTCCGGAAATGTGTGTCAGAATACAAGAGGAAGCTACCACTGTCACAAGATCGAGTGCCCGACTGGATATAGACTAGAGTCTAAGCACCGATGCACAAGAATACAGAGAACATGCCTGGTGTCAGATTGGTCGTGTCTGCAATTGCCCAGTACATACAGCTATCACTTCATAACGTTCGTCGCAAACATATTCCTGCCTTCGGGAAGCGTGGACCTATTCACGATGCACGGTCCTTCGTGGCAAGACTCTGTGGTCAGCTTCGAAATGAGAATGATAAGCGTTCAAGCCTCACACGGAGTGCAACCAACAGATTTGCGATGTTTCGACATGCGTCCGTCGGGCAATATCTGCGTAGTGTCGCTATTGTGCTCACTGGGAGGCCCTCAAGTGGCTGAATTAGAACTGACGATGTCCCTGTACCAAAGGAGCCAGTTCGCGGGCAGCGCTGTCGCGAGATTAGTTGTCATTGTATCTGAATACGAATATTAG
Protein
MKFIVVIFTFLFSTVNALSSEETLAVTDSCCTHAEGAALLSTDCSAIQPPEDIDAEQLPTCITALKACCEKQHKEKDECSAGVKLASSKRCNSIATNIGQACCEECSIGRTIGKTQGLEGCGTPVNEEYNSPSTILRKKAFHQCCSAAANEPETTEKVETTTLPSTTTSTAKPKVKCTKDSCAQICEVEQETIRCSCRDGFRLHSDKKSCKDVNECAEADDDICTEEGFVCHNTPGSYKCVPLKPTTKRDVGSNCPPGFKKNVLNQVCDDINECQLPRPPCPKYLCENTIGGYKCAGKPGKPVIESAPGAPEVTTPVAPVRNEICPPGFRAGPDDECIDIDECEESLDDCQRLSQHCINTHGSFFCQDHVSKRCAPGFKVNTSTGICEDIDECEESTEVCKRTEICVNLPGAYNCKSKISSLPKLAKTKCQEGMRLRPGGTVCEDIDECREGTHLCDEFQNCINTYGAHECRCKNGFELDSLLESCVDIDECALKLDNCGPRLHCLNVLGSFTCTYRQPSTTSTTPVPPLYEYEYYDSEDDNETAPEIAATTSTTRPTYPTQRTTTSTQRTTTSTQRTTTSTPRTTTSTQRTTTSTTKPTTLTTRRSRPYYPYYDRPRYTTSTKRPSYPYNRRPSTPSRVEPTTPKKPEVTSSTEVSRRPDYNPSRSDYTPRRPEITTPIKPETTTEVFRRPTHSPSRPDNSRRPSPFRPGYVTGMTRPKDPITIEKEREDDGGIELDTNTIPKETWTNVIGRDKPEATDTPAFDPNEVHCLNGYEKDFFGNCVDINECERSHICSNLEDCINKAGGYRCDCIAGFKRNQQGWCIPIPPPTTTSTTSTTTSTTTPRTISVSLPEWQYTPPRSRGRYPSIACELGYTYDTKYGKCVDIDECATNKATCSRNEECVNIDGGFRCVCGWRCRAENEKPTADPSLVPASPISPIAPHPPPPSYSPSEPRYPDNTNVITVGAQYGQRGPWSLRPSFTRFSDRGAVPCSSYSRCKNTVGSYRCECDEGFRNAASNDKVCVDVDECSESPNVCQHGCANTWGGYRCYCKRGYRVGNDNRTCVDVNECEEWTSVRIRGKLCAGECVNEPGSYRCACPAGYRLSEDNRSCIDIDECETGEAPCARTSDSGNVCQNTRGSYHCHKIECPTGYRLESKHRCTRIQRTCLVSDWSCLQLPSTYSYHFITFVANIFLPSGSVDLFTMHGPSWQDSVVSFEMRMISVQASHGVQPTDLRCFDMRPSGNICVVSLLCSLGGPQVAELELTMSLYQRSQFAGSAVARLVVIVSEYEY
Summary
Uniprot
ProteinModelPortal
PDB
3V65
E-value=1.05508e-10,
Score=165
Ontologies
GO
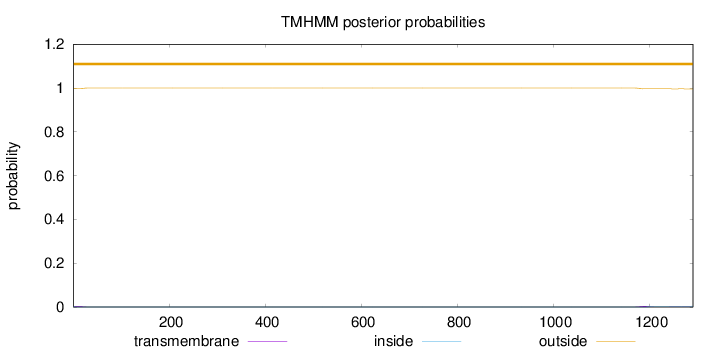
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.999616
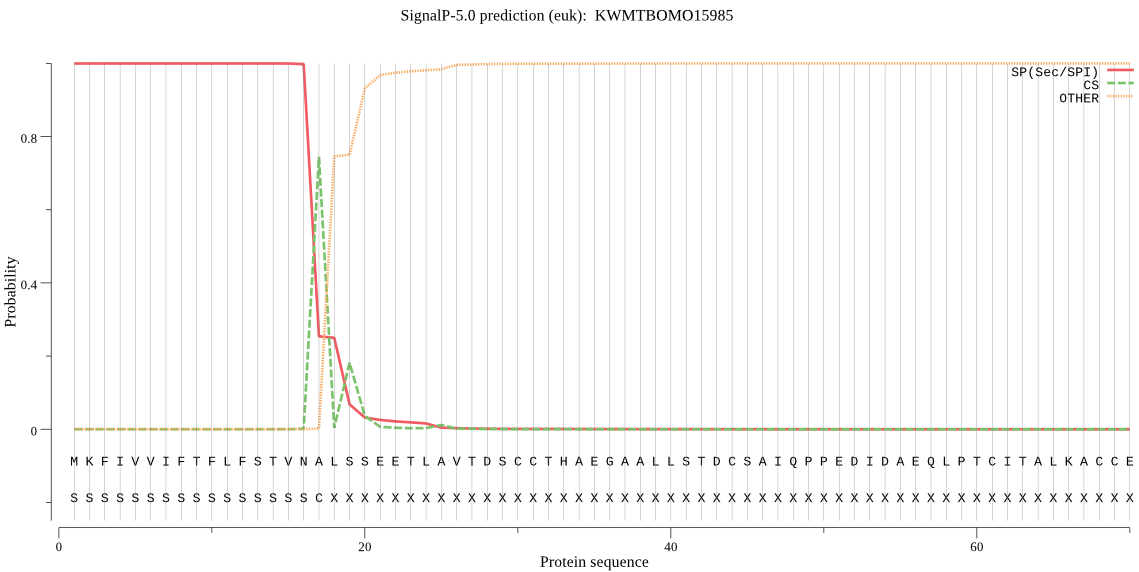
Length:
1291
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.14704
Exp number, first 60 AAs:
0.05593
Total prob of N-in:
0.00297
outside
1 - 1291
Population Genetic Test Statistics
Pi
209.684022
Theta
160.010064
Tajima's D
0.879846
CLR
0.097012
CSRT
0.623868806559672
Interpretation
Uncertain
