Pre Gene Modal
BGIBMGA005735
Annotation
PREDICTED:_delta(24)-sterol_reductase-like_isoform_X1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.862
Sequence
CDS
ATGGCCATAGAAACAGAAACGTTTTTGGAGTACCTCGTAGTGGAATATCGATGGGTGATAGTAATATTGGCCCTGTTGCCTATGTCAGCGGCATGGAAGTTATGGTCGATAATCAGGAACTACGTTGTGTTCAAGATGAATAGTGCTCCAAAGATGCATGATGATAAAGTCAAAGAGGTTCAAAGACAGATAAAAGAATGGTTATCCGGGGACAAGTCGACTCATCTCTGCACAGCCCGGCCAACATGGCAAACAATGTCTTTTCGACACAGCATGTACAAGAGGACATTTACCAACATACAAATTAATCTAGTCGATGTCTTGGAGGTGGACAAAGAGAATATGACAGTCCGTTGCGAGCCTCTAGTGACGATGGGTCAGCTGTCCCGCACATTGGCGCCGCTAGGCTTGGCGCTGCCCGTTGTTCCAGAGTTAGACCAACTCACAGTCGGCGGCCTGGTGATGGGGACCGGCGTGGAGACAACATCGCACGTCCACGGGCTTTTTCAGCATGTGTGCCTGGAGTACGAGCTGGTTCTCGCCGACGGCTCTGTCGTTAACTGTAGTAAGGACGAAAACGCTGATCTATTTTACGCTGTTCCGTGGTCTTACGGCACCCTTGGATTTCTGACTTCAGTTGTGATCAAAGTTATTCCGGCTAAGAAGTATGTCCGCATACATTATTACCCTTTCAACAATCTTTCCGACCTAGCAAAGCGGTTCAGTCAAGAATCTTTGAAGCCGAACCCTCACCAATTCGTTGAGGCCTTGCTTTACACTAGAGACTCGGGCGTGCTCATGTTAGGAGACATGGTGGATGAAGTCGGCAGAGATGGAAAGCTCAATCCAATAGGGGTGTGGCACGCGGAATGGTTCTTCAAACAAGTGGAGAAGCATTTAAAAAGAAAGCGCACCGCGATAGAGTATATACCTCTTAGAGATTACTACCATCGTCACACTAGAAGTCTGTTCTGGGAATTGCAAGACATAATTTCGTTTGGGAACAACTTTATTTTCCGCTACCTGTTCGGCTGGTTGATGCCTCCGGAGGTCTCCTTATTGAAGTTGACGCAGCCCGAAGCCGTCACCAAGTTGTACAACAAGGCTCACGTGATACAAGACATGCTAATACCAATTGAGCTCTTGGAGAAAGCGATTGCGTTCTTTCACGACGAATTCGAGGTATACCCTATATGGTTGTGTCCTTTCAAAATATTCAACAACCCTGGACAGCTGAAGATAAAACCTGGCGAGGAATCACAGATGTTTGTCGATATCGGAGTGTATGGCGTTCCAAAAGCCAAAGGATTTGAAACAATTGCATCGACGCGTCACGTAGAGAGCTTCGTGATCCAGAACCAAGGCTTCCAAATGTTATATGCCGATACCTACACAACGAGGGAGGAATTCAGACAAATGTTCGACCATAAACTTTACGACAGAGTGCGGGCTTCGCTGCCTAATTGCAAGGAAGCCTTCCCCGAGATCTATGATAAAGTCAACAGAAATGTTAGGAAATAA
Protein
MAIETETFLEYLVVEYRWVIVILALLPMSAAWKLWSIIRNYVVFKMNSAPKMHDDKVKEVQRQIKEWLSGDKSTHLCTARPTWQTMSFRHSMYKRTFTNIQINLVDVLEVDKENMTVRCEPLVTMGQLSRTLAPLGLALPVVPELDQLTVGGLVMGTGVETTSHVHGLFQHVCLEYELVLADGSVVNCSKDENADLFYAVPWSYGTLGFLTSVVIKVIPAKKYVRIHYYPFNNLSDLAKRFSQESLKPNPHQFVEALLYTRDSGVLMLGDMVDEVGRDGKLNPIGVWHAEWFFKQVEKHLKRKRTAIEYIPLRDYYHRHTRSLFWELQDIISFGNNFIFRYLFGWLMPPEVSLLKLTQPEAVTKLYNKAHVIQDMLIPIELLEKAIAFFHDEFEVYPIWLCPFKIFNNPGQLKIKPGEESQMFVDIGVYGVPKAKGFETIASTRHVESFVIQNQGFQMLYADTYTTREEFRQMFDHKLYDRVRASLPNCKEAFPEIYDKVNRNVRK
Summary
Uniprot
H9J893
A0A2H1VSR8
A0A2A4JI87
A0A212FJA8
A0A194Q530
A0A2H1VSW5
+ More
A0A3R7Q404 A0A1D2MFS4 A0A2P2HY56 A0A210QJU8 A0A139WPD9 Q16MZ3 A0A1S3JWJ9 A0A1S4FVR0 B0W0K1 A0A067RPY2 A0A1B0D7D2 V5GS72 H3ACL2 C7DR08 A0A1L8DZ79 A0A1L8DZB4 V4BLP6 A0A0P5V4A0 A0A0P5Z7T1 R7UR55 U4UWW1 A0A0P5KFF5 K7F6B9 J3JWF2 A0A2J7PP71 M7B6K0 A0A151M4G4 A0A218UXF9 A0A0P6FTL8 A0A0P5LYF3 A0A0P6G5Y6 A0A3M0KCB9 A0A0B8RP34 A0A0P6F118 A0A1V4JZG3 A0A0N8B4W4 A0A226PQZ4 A0A226NBH2 L9L3P5 Q5ZIF2 A0A3B3RB58 T0SCN5 A0A1W4WSS7 A0A0N8ECR9 A0A3L8SQF0 A0A1V9YJ41 A0A0P6G0D2 K1R4Q8 H2UXJ0 A0A0P5R8M6 A0A0P6FKA2 A0A3Q7UXW5 G3PAV7 A0A3Q3VSK1 A0A3B5ADT0 J3S482 G3PAW7 A0A267GHJ0 A0A1W7RD14 A0A267ECF4 T1E428 A0A0B7AT95 A0A0Q3PK92 F1QUM4 U3KCC1 M3Y6T6 Q5PRC9 A0A337STK0 A0A2H6N2F6 A0A024UF76 A0A2U4BXN9 F6YWP5 A0A3Q3F179 A0A2Y9DCD2 D2H2H4 G1NFY0 A0A0P6J636 F1PXA2 A0A3Q7TN91 I3J3V6 A0A2K6JRZ4 A0A2Y9GKA1 A0A2U3VY51 A0A2Y9IY12 K9IKX2 A0A3Q7P1Q8 A0A341AX57 A0A2Y9N9K0 A0A2Y9EX06 A0A3Q1B0U7 A0A340YGL2 G3R5L2 A0A2I2YB07 A0A2K6EY76
A0A3R7Q404 A0A1D2MFS4 A0A2P2HY56 A0A210QJU8 A0A139WPD9 Q16MZ3 A0A1S3JWJ9 A0A1S4FVR0 B0W0K1 A0A067RPY2 A0A1B0D7D2 V5GS72 H3ACL2 C7DR08 A0A1L8DZ79 A0A1L8DZB4 V4BLP6 A0A0P5V4A0 A0A0P5Z7T1 R7UR55 U4UWW1 A0A0P5KFF5 K7F6B9 J3JWF2 A0A2J7PP71 M7B6K0 A0A151M4G4 A0A218UXF9 A0A0P6FTL8 A0A0P5LYF3 A0A0P6G5Y6 A0A3M0KCB9 A0A0B8RP34 A0A0P6F118 A0A1V4JZG3 A0A0N8B4W4 A0A226PQZ4 A0A226NBH2 L9L3P5 Q5ZIF2 A0A3B3RB58 T0SCN5 A0A1W4WSS7 A0A0N8ECR9 A0A3L8SQF0 A0A1V9YJ41 A0A0P6G0D2 K1R4Q8 H2UXJ0 A0A0P5R8M6 A0A0P6FKA2 A0A3Q7UXW5 G3PAV7 A0A3Q3VSK1 A0A3B5ADT0 J3S482 G3PAW7 A0A267GHJ0 A0A1W7RD14 A0A267ECF4 T1E428 A0A0B7AT95 A0A0Q3PK92 F1QUM4 U3KCC1 M3Y6T6 Q5PRC9 A0A337STK0 A0A2H6N2F6 A0A024UF76 A0A2U4BXN9 F6YWP5 A0A3Q3F179 A0A2Y9DCD2 D2H2H4 G1NFY0 A0A0P6J636 F1PXA2 A0A3Q7TN91 I3J3V6 A0A2K6JRZ4 A0A2Y9GKA1 A0A2U3VY51 A0A2Y9IY12 K9IKX2 A0A3Q7P1Q8 A0A341AX57 A0A2Y9N9K0 A0A2Y9EX06 A0A3Q1B0U7 A0A340YGL2 G3R5L2 A0A2I2YB07 A0A2K6EY76
Pubmed
19121390
22118469
26354079
27289101
28812685
18362917
+ More
19820115 17510324 24845553 9215903 19727923 21292972 23254933 23537049 17381049 22516182 23624526 22293439 25476704 24621616 23385571 15592404 15642098 29240929 30282656 25527045 22992520 21551351 23025625 26358130 23758969 23594743 17975172 12481130 15114417 20010809 20838655 16341006 25186727 22398555
19820115 17510324 24845553 9215903 19727923 21292972 23254933 23537049 17381049 22516182 23624526 22293439 25476704 24621616 23385571 15592404 15642098 29240929 30282656 25527045 22992520 21551351 23025625 26358130 23758969 23594743 17975172 12481130 15114417 20010809 20838655 16341006 25186727 22398555
EMBL
BABH01038484
BABH01038485
BABH01038486
ODYU01004219
SOQ43867.1
NWSH01001391
+ More
PCG71428.1 AGBW02008280 OWR53837.1 KQ459460 KPJ00647.1 ODYU01004218 SOQ43866.1 QCYY01002735 ROT67970.1 LJIJ01001403 ODM91838.1 IACF01000970 LAB66703.1 NEDP02003363 OWF48891.1 KQ971307 KYB29727.1 CH477844 EAT35716.1 DS231818 EDS41303.1 KK852539 KDR21794.1 AJVK01026864 GALX01004024 JAB64442.1 AFYH01161974 AFYH01161975 GQ199604 GL732536 ACT20729.1 EFX83687.1 GFDF01002316 JAV11768.1 GFDF01002317 JAV11767.1 KB202567 ESO89669.1 GDIP01104995 JAL98719.1 GDIP01215787 GDIP01048802 LRGB01001409 JAM54913.1 KZS12025.1 AMQN01007658 KB301107 ELU05906.1 KB632399 ERL94765.1 GDIQ01250843 GDIQ01210640 JAK41085.1 AGCU01155374 AGCU01155375 AGCU01155376 BT127570 AEE62532.1 NEVH01023279 PNF18142.1 KB535465 EMP33601.1 AKHW03006631 KYO19415.1 MUZQ01000109 OWK58100.1 GDIQ01222300 GDIQ01212546 GDIQ01204799 GDIQ01055763 GDIQ01049573 GDIQ01044563 JAN50174.1 GDIQ01172409 JAK79316.1 GDIQ01039517 JAN55220.1 QRBI01000117 RMC08720.1 GBSH01002627 JAG66399.1 GDIQ01055818 JAN38919.1 LSYS01005497 OPJ77037.1 GDIQ01212547 JAK39178.1 AWGT02000029 OXB82114.1 MCFN01000116 OXB64570.1 KB320587 ELW67987.1 AADN05000647 AJ720832 CAG32491.1 JH767136 EQC40607.1 GDIQ01039516 JAN55221.1 QUSF01000010 RLW05965.1 JNBR01001604 OQR85741.1 GDIQ01058462 JAN36275.1 JH818314 EKC40753.1 GDIQ01111090 JAL40636.1 GDIQ01058463 JAN36274.1 JU174311 GBEX01003298 AFJ49837.1 JAI11262.1 NIVC01001081 NIVC01000326 PAA72549.1 PAA85466.1 GDAY02002401 JAV49031.1 NIVC01002298 PAA59076.1 GAAZ01002425 JAA95518.1 HACG01036967 CEK83832.1 LMAW01002834 KQK76604.1 BX511260 AGTO01000977 AEYP01029325 AEYP01029326 BC086711 BC165211 AAH86711.1 AAI65211.1 AANG04003279 IACI01032420 LAA23103.1 KI913957 ETW04537.1 EAAA01002575 ACTA01074127 ACTA01082127 ACTA01090127 GL192442 EFB29172.1 GEBF01003918 JAN99714.1 AAEX03003811 AERX01004372 GABZ01005585 JAA47940.1 CABD030003368 CABD030003369
PCG71428.1 AGBW02008280 OWR53837.1 KQ459460 KPJ00647.1 ODYU01004218 SOQ43866.1 QCYY01002735 ROT67970.1 LJIJ01001403 ODM91838.1 IACF01000970 LAB66703.1 NEDP02003363 OWF48891.1 KQ971307 KYB29727.1 CH477844 EAT35716.1 DS231818 EDS41303.1 KK852539 KDR21794.1 AJVK01026864 GALX01004024 JAB64442.1 AFYH01161974 AFYH01161975 GQ199604 GL732536 ACT20729.1 EFX83687.1 GFDF01002316 JAV11768.1 GFDF01002317 JAV11767.1 KB202567 ESO89669.1 GDIP01104995 JAL98719.1 GDIP01215787 GDIP01048802 LRGB01001409 JAM54913.1 KZS12025.1 AMQN01007658 KB301107 ELU05906.1 KB632399 ERL94765.1 GDIQ01250843 GDIQ01210640 JAK41085.1 AGCU01155374 AGCU01155375 AGCU01155376 BT127570 AEE62532.1 NEVH01023279 PNF18142.1 KB535465 EMP33601.1 AKHW03006631 KYO19415.1 MUZQ01000109 OWK58100.1 GDIQ01222300 GDIQ01212546 GDIQ01204799 GDIQ01055763 GDIQ01049573 GDIQ01044563 JAN50174.1 GDIQ01172409 JAK79316.1 GDIQ01039517 JAN55220.1 QRBI01000117 RMC08720.1 GBSH01002627 JAG66399.1 GDIQ01055818 JAN38919.1 LSYS01005497 OPJ77037.1 GDIQ01212547 JAK39178.1 AWGT02000029 OXB82114.1 MCFN01000116 OXB64570.1 KB320587 ELW67987.1 AADN05000647 AJ720832 CAG32491.1 JH767136 EQC40607.1 GDIQ01039516 JAN55221.1 QUSF01000010 RLW05965.1 JNBR01001604 OQR85741.1 GDIQ01058462 JAN36275.1 JH818314 EKC40753.1 GDIQ01111090 JAL40636.1 GDIQ01058463 JAN36274.1 JU174311 GBEX01003298 AFJ49837.1 JAI11262.1 NIVC01001081 NIVC01000326 PAA72549.1 PAA85466.1 GDAY02002401 JAV49031.1 NIVC01002298 PAA59076.1 GAAZ01002425 JAA95518.1 HACG01036967 CEK83832.1 LMAW01002834 KQK76604.1 BX511260 AGTO01000977 AEYP01029325 AEYP01029326 BC086711 BC165211 AAH86711.1 AAI65211.1 AANG04003279 IACI01032420 LAA23103.1 KI913957 ETW04537.1 EAAA01002575 ACTA01074127 ACTA01082127 ACTA01090127 GL192442 EFB29172.1 GEBF01003918 JAN99714.1 AAEX03003811 AERX01004372 GABZ01005585 JAA47940.1 CABD030003368 CABD030003369
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000283509
UP000094527
+ More
UP000242188 UP000007266 UP000008820 UP000085678 UP000002320 UP000027135 UP000092462 UP000008672 UP000000305 UP000030746 UP000076858 UP000014760 UP000030742 UP000007267 UP000235965 UP000031443 UP000050525 UP000197619 UP000269221 UP000190648 UP000198419 UP000198323 UP000011518 UP000000539 UP000261540 UP000030762 UP000192223 UP000276834 UP000243579 UP000005408 UP000005226 UP000286642 UP000007635 UP000261620 UP000261400 UP000215902 UP000051836 UP000000437 UP000016665 UP000000715 UP000011712 UP000024375 UP000245320 UP000008144 UP000261660 UP000248480 UP000008912 UP000001645 UP000002254 UP000286640 UP000005207 UP000233180 UP000248481 UP000245340 UP000248482 UP000286641 UP000252040 UP000248483 UP000248484 UP000257160 UP000265300 UP000001519 UP000233160
UP000242188 UP000007266 UP000008820 UP000085678 UP000002320 UP000027135 UP000092462 UP000008672 UP000000305 UP000030746 UP000076858 UP000014760 UP000030742 UP000007267 UP000235965 UP000031443 UP000050525 UP000197619 UP000269221 UP000190648 UP000198419 UP000198323 UP000011518 UP000000539 UP000261540 UP000030762 UP000192223 UP000276834 UP000243579 UP000005408 UP000005226 UP000286642 UP000007635 UP000261620 UP000261400 UP000215902 UP000051836 UP000000437 UP000016665 UP000000715 UP000011712 UP000024375 UP000245320 UP000008144 UP000261660 UP000248480 UP000008912 UP000001645 UP000002254 UP000286640 UP000005207 UP000233180 UP000248481 UP000245340 UP000248482 UP000286641 UP000252040 UP000248483 UP000248484 UP000257160 UP000265300 UP000001519 UP000233160
PRIDE
Interpro
IPR016166
FAD-bd_PCMH
+ More
IPR040165 Diminuto-like
IPR016169 FAD-bd_PCMH_sub2
IPR006094 Oxid_FAD_bind_N
IPR036318 FAD-bd_PCMH-like_sf
IPR016164 FAD-linked_Oxase-like_C
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR000742 EGF-like_dom
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR026823 cEGF
IPR009030 Growth_fac_rcpt_cys_sf
IPR001881 EGF-like_Ca-bd_dom
IPR040165 Diminuto-like
IPR016169 FAD-bd_PCMH_sub2
IPR006094 Oxid_FAD_bind_N
IPR036318 FAD-bd_PCMH-like_sf
IPR016164 FAD-linked_Oxase-like_C
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR000742 EGF-like_dom
IPR018097 EGF_Ca-bd_CS
IPR013032 EGF-like_CS
IPR026823 cEGF
IPR009030 Growth_fac_rcpt_cys_sf
IPR001881 EGF-like_Ca-bd_dom
Gene 3D
ProteinModelPortal
H9J893
A0A2H1VSR8
A0A2A4JI87
A0A212FJA8
A0A194Q530
A0A2H1VSW5
+ More
A0A3R7Q404 A0A1D2MFS4 A0A2P2HY56 A0A210QJU8 A0A139WPD9 Q16MZ3 A0A1S3JWJ9 A0A1S4FVR0 B0W0K1 A0A067RPY2 A0A1B0D7D2 V5GS72 H3ACL2 C7DR08 A0A1L8DZ79 A0A1L8DZB4 V4BLP6 A0A0P5V4A0 A0A0P5Z7T1 R7UR55 U4UWW1 A0A0P5KFF5 K7F6B9 J3JWF2 A0A2J7PP71 M7B6K0 A0A151M4G4 A0A218UXF9 A0A0P6FTL8 A0A0P5LYF3 A0A0P6G5Y6 A0A3M0KCB9 A0A0B8RP34 A0A0P6F118 A0A1V4JZG3 A0A0N8B4W4 A0A226PQZ4 A0A226NBH2 L9L3P5 Q5ZIF2 A0A3B3RB58 T0SCN5 A0A1W4WSS7 A0A0N8ECR9 A0A3L8SQF0 A0A1V9YJ41 A0A0P6G0D2 K1R4Q8 H2UXJ0 A0A0P5R8M6 A0A0P6FKA2 A0A3Q7UXW5 G3PAV7 A0A3Q3VSK1 A0A3B5ADT0 J3S482 G3PAW7 A0A267GHJ0 A0A1W7RD14 A0A267ECF4 T1E428 A0A0B7AT95 A0A0Q3PK92 F1QUM4 U3KCC1 M3Y6T6 Q5PRC9 A0A337STK0 A0A2H6N2F6 A0A024UF76 A0A2U4BXN9 F6YWP5 A0A3Q3F179 A0A2Y9DCD2 D2H2H4 G1NFY0 A0A0P6J636 F1PXA2 A0A3Q7TN91 I3J3V6 A0A2K6JRZ4 A0A2Y9GKA1 A0A2U3VY51 A0A2Y9IY12 K9IKX2 A0A3Q7P1Q8 A0A341AX57 A0A2Y9N9K0 A0A2Y9EX06 A0A3Q1B0U7 A0A340YGL2 G3R5L2 A0A2I2YB07 A0A2K6EY76
A0A3R7Q404 A0A1D2MFS4 A0A2P2HY56 A0A210QJU8 A0A139WPD9 Q16MZ3 A0A1S3JWJ9 A0A1S4FVR0 B0W0K1 A0A067RPY2 A0A1B0D7D2 V5GS72 H3ACL2 C7DR08 A0A1L8DZ79 A0A1L8DZB4 V4BLP6 A0A0P5V4A0 A0A0P5Z7T1 R7UR55 U4UWW1 A0A0P5KFF5 K7F6B9 J3JWF2 A0A2J7PP71 M7B6K0 A0A151M4G4 A0A218UXF9 A0A0P6FTL8 A0A0P5LYF3 A0A0P6G5Y6 A0A3M0KCB9 A0A0B8RP34 A0A0P6F118 A0A1V4JZG3 A0A0N8B4W4 A0A226PQZ4 A0A226NBH2 L9L3P5 Q5ZIF2 A0A3B3RB58 T0SCN5 A0A1W4WSS7 A0A0N8ECR9 A0A3L8SQF0 A0A1V9YJ41 A0A0P6G0D2 K1R4Q8 H2UXJ0 A0A0P5R8M6 A0A0P6FKA2 A0A3Q7UXW5 G3PAV7 A0A3Q3VSK1 A0A3B5ADT0 J3S482 G3PAW7 A0A267GHJ0 A0A1W7RD14 A0A267ECF4 T1E428 A0A0B7AT95 A0A0Q3PK92 F1QUM4 U3KCC1 M3Y6T6 Q5PRC9 A0A337STK0 A0A2H6N2F6 A0A024UF76 A0A2U4BXN9 F6YWP5 A0A3Q3F179 A0A2Y9DCD2 D2H2H4 G1NFY0 A0A0P6J636 F1PXA2 A0A3Q7TN91 I3J3V6 A0A2K6JRZ4 A0A2Y9GKA1 A0A2U3VY51 A0A2Y9IY12 K9IKX2 A0A3Q7P1Q8 A0A341AX57 A0A2Y9N9K0 A0A2Y9EX06 A0A3Q1B0U7 A0A340YGL2 G3R5L2 A0A2I2YB07 A0A2K6EY76
PDB
4MLA
E-value=2.69854e-05,
Score=115
Ontologies
PATHWAY
GO
GO:0016491
GO:0016021
GO:0071949
GO:0005509
GO:0008202
GO:0016628
GO:0016020
GO:0005737
GO:0000246
GO:0006695
GO:0042987
GO:0061024
GO:0008104
GO:0042605
GO:0043154
GO:0005634
GO:0043588
GO:0006979
GO:0005783
GO:0031639
GO:0030539
GO:0009888
GO:0019899
GO:0050660
GO:0003824
GO:0016614
GO:0004888
GO:0007166
GO:0000287
GO:0006281
GO:0006259
GO:0055114
PANTHER
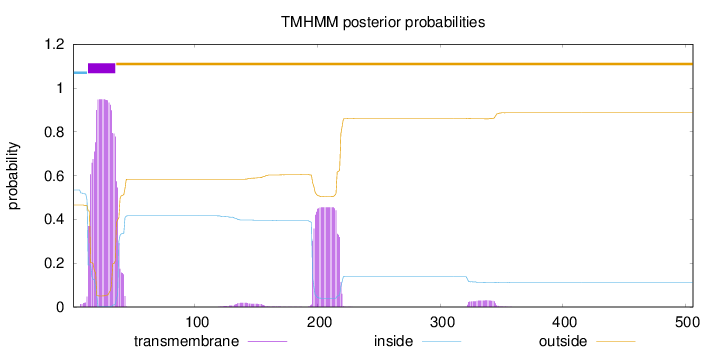
Topology
Length:
506
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
32.01067
Exp number, first 60 AAs:
20.91548
Total prob of N-in:
0.53427
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 506
Population Genetic Test Statistics
Pi
157.792367
Theta
158.242755
Tajima's D
0.386419
CLR
0.318346
CSRT
0.473226338683066
Interpretation
Uncertain
